Abstract
In the cyanobacteria Synechococcus elongatus and Thermosynechococcus elongatus, the KaiA, KaiB and KaiC proteins in the presence of ATP generate a post-translational oscillator (PTO) that can be reconstituted in vitro. KaiC is the result of a gene duplication and resembles a double doughnut with N-terminal CI and C-terminal CII hexameric rings. Six ATPs are bound between subunits in both the CI and CII ring. CI harbors ATPase activity, and CII catalyzes phosphorylation and dephosphorylation at T432 and S431 with a ca. 24-h period. KaiA stimulates KaiC phosphorylation, and KaiB promotes KaiC subunit exchange and sequesters KaiA on the KaiB-KaiC interface in the final stage of the clock cycle. Studies of the PTO protein-protein interactions are convergent in terms of KaiA binding to CII but have led to two opposing models of the KaiB-KaiC interaction. Electron microscopy (EM) and small angle X-ray scattering (SAXS), together with native PAGE using full-length proteins and separate CI and CII rings, are consistent with binding of KaiB to CII. Conversely, NMR together with gel filtration chromatography and denatured PAGE using monomeric CI and CII domains support KaiB binding to CI. To resolve the existing controversy, we studied complexes between KaiB and gold-labeled, full-length KaiC with negative stain EM. The EM data clearly demonstrate that KaiB contacts the CII ring. Together with the outcomes of previous analyses, our work establishes that only CII participates in interactions with KaiA and KaiB as well as with the His kinase SasA involved in the clock output pathway.
Introduction
The circadian clock in the cyanobacteria Synechococcus elongatus (S. elongatus) and Thermosynechococcus elongatus (T. elongatus) is composed of an inner, post-translational oscillator (PTO) that is coupled to a transcription-translation oscillator feedback loop (TTFL) and controls gene expression in these cyanobacteria genome-wide in a non-promoter-specific fashion.Citation1,Citation2 The inner timer is constituted of three proteins, KaiA, KaiB and KaiC, that produce a daily cycle of KaiC phosphorylation and dephosphorylation. Remarkably, the PTO can be reconstituted in vitro from the three proteins in the presence of ATP to tick in a temperature-compensated manner with a period of ca. 24 h.Citation3 Its relative simplicity renders this system an ideal target for a detailed biochemical and biophysical characterization of the cogs and gears underlying a circadian clock.Citation4
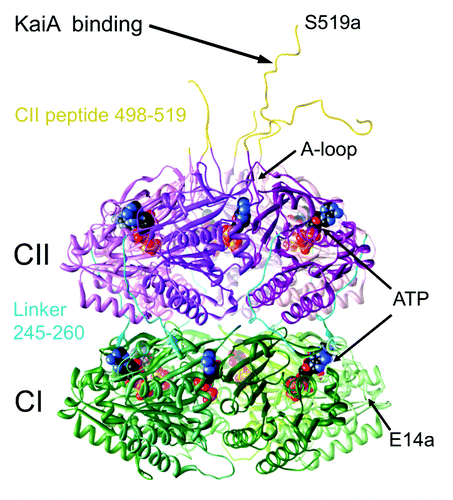
KaiC is the central component of the clock and is the result of a gene duplication.Citation5,Citation6 Both halves contain Walker A and B motifs associated with ATP binding and processing and the protein constitutes a homo-hexamer.Citation7 The three-dimensional structure of KaiC from S. elongatus was determined by X-ray crystallography and revealed two stacked rings linked by single-peptide chains of 15 amino acids between the domains assembling the N-terminal CI and C-terminal CII rings ().Citation8 Twelve ATP molecules are lodged between subunits in the CI and CII rings. The CI ring catalyzes ATP hydrolysis, and the CII ring exhibits auto-phosphorylation and auto-dephosphorylation activities that target residues T432 and S431, whereby phosphorylation takes place across the subunit interface.Citation9 Rather than separate kinase and phosphatase active sites, KaiC uses the same site at the subunit interface to catalyze both reactions, whereby dephosphorylation proceeds via a phospho-transferase (ATP synthase) mechanism.Citation10,Citation11 Phosphorylation and dephosphorylation of T432 and S431 follow a strict order, such that the threonine receives a phosphate group first and loses it first, following a hyper-phosphorylated state during which threonine and serine both carry a phosphate.Citation12,Citation13 KaiA stimulates KaiC phosphorylation by binding to a portion of the C-terminal CII peptide and unraveling the so-called A-loop (),Citation14,Citation15 an event that triggers phosphorylation of all six subunits in a concerted allosteric fashion.Citation16 Whereas KaiA can be clearly mapped to the C-terminal end of the CII ring, establishing the binding mode of KaiB to KaiC has turned out to be more complicated.
Figure 1. Crystal structure of the S. elongatus KaiC hexamer (PDB ID 3DVL).Citation15
Two investigations arrived at the conclusion that KaiB binds KaiC on the same side as KaiA, i.e., via CII. Akiyama and coworkers used small angle X-ray scattering (SAXS) to generate 3D envelopes of binary KaiAC and KaiBC complexes.Citation17 A hypothetical model of the ternary KaiABC complex was generated by superimposing the two binary models and pasting the KaiB tetramer on top of KaiCII, either in close vicinity of N-terminal pseudoreceiver domains from KaiA dimer or C-terminal α-helical bundle domains attached to the KaiCII peptide. The authors’ choice of CII was likely influenced by the fact that KaiB antagonizes KaiA action,Citation18 possibly by competing for regions on CII with some degree of overlap. We used separate CI and CII rings from S. elongatus in combination with native PAGE to establish that KaiB does not bind to CI hexamer but produces a bandshift when mixed with CII.Citation19 However, negative stain EM revealed that whereas CI exists predominantly in the hexameric form in the presence of ATP, CII does not, consistent with the higher rigidity of KaiCICitation20 and more tightly bound ATP in that half.Citation8 A negative stain and cryo EM analysis of the KaiBC complex revealed that KaiB bound as two dimers on top of CII (the ring identified by native PAGE).Citation19 That KaiB can exist in the dimeric form and is able to generate normal circadian oscillations in the KaiABC in vitro system was later confirmed independently.Citation21
A recent study based on solution NMR and using monomeric CI and CII domains identified the KaiB binding site on CI and argued that this site is hidden when CI is in the hexameric form.Citation22 However, stacking among the CII and CI rings and wedging apart of subunits by ADP exposes the site and results in docking of KaiB and dephosphorylation of KaiC by sequestration of KaiA.Citation23-Citation25 These two opposing binding modes, KaiB interacting with CII vs. KaiB interacting with CI, cannot be reconciled in a single mechanistic model of how KaiB recognizes the phosphorylated state of KaiC, i.e., the preferred TpS431 form,Citation12,Citation26 and subsequently promotes KaiC subunit exchangeCitation27,Citation28 and mediates dephosphorylation by sequestering KaiA. One can readily explain specific binding of KaiB to the phosphorylated form based on the model that has KaiB bound to the CII ring, because the electrostatic surface potential (ESP) of that ring is altered upon phosphorylation of T432 and S431.Citation25 KaiB loosens the interactions among subunits from the intrinsically weaker side of the KaiC hexamer and thus promotes subunit exchange. Formation of the KaiBC interface leads to sequestration of KaiA, the latter either bound on the CII surface in close vicinity from KaiB or existing in the free form, resulting in a ternary KaiABC complex of distinct shapeCitation25,Citation28 and dephosphorylation of KaiC. In the alternative model, with KaiB contacting KaiC from the CI side, stacking between the CII and CI rings would result in splaying of CI domains and binding of KaiB to a site previously hidden at the subunit interface.Citation22 The newly formed KaiBC complex would then promote desphosphorylation of KaiC by sequestering KaiA. It is important to remember in this context that the CI and CII rings are stacked in the crystal structure,Citation8 and that molecular envelopes of the KaiBC complex based on either EMCitation19 or SAXSCitation25 do not exhibit an asymmetry of the two rings in terms of their widths. Moreover, although we relied on oligomers of separate CI and CII domains to determine the identity of the ring recognized by KaiB,Citation19 the EM analysis was conducted with full-length KaiC, thus precluding potential artifacts that might be caused by the use of partial protein constructs.
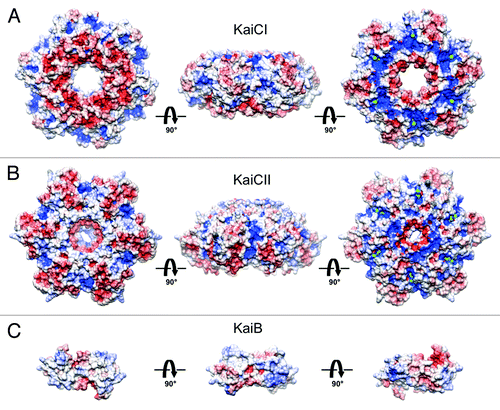
The CI and CII rings of KaiC exhibit distinct shapes and ESPs. The disk-shaped CI ring features similar curvatures on the upper (buried at the CI-CII interface) and lower sides (). In contrast, the CII ring is slightly thicker, and its C-terminal surface is dome-shaped (), with peptides that comprise 30 amino acids each sprouting from the central region (). These distinct shapes and the opposing polarities of the ESPs of CI at the bottom of KaiC and CII at the top of KaiC () virtually preclude that KaiB can bind to both. To definitively settle the question of whether KaiB binds to CI or CII, and relying only on the physiologically relevant, full-length form of KaiC preferred by KaiB (the S431D mutant, a mimic of the pS431 phosphorylation stateCitation12), we conducted a negative-stain EM analysis of the KaiBC complex with gold-labeled KaiC. The EM data are consistent with binding of KaiB to CII and the presence of two KaiB dimers on top of KaiC.Citation19 Because KaiB and SasA compete for similar binding sites on KaiC,Citation25 our work also affirms SasA binding to the CII ringCitation25 and not to CI.Citation29,Citation30 It also follows that KaiBC sequesters KaiA on the CII side, consistent with the previously presented EM model of the ternary KaiABC complex.Citation25
Figure 2. Electrostatic surface potentials (ESPs) of individual rings in S. elongatus KaiC (PDB ID 3DVL) and T. elongatus KaiB (PDB ID 2QKE).Citation19 ESP of (A) the CI ring, (B) the CII ring and (C) KaiB. KaiC rings are viewed from the N terminus, the side and the C terminus (from left to right). KaiB dimer is viewed along the normal to the non-crystallographic dyad (center) and rotated by −90° (left) and +90° (right) around the horizontal axis. The minimum and maximum values of the electrostatic potential are −5 and +5 kT/e, respectively. ESPs were calculated using default parameters of APBS.Citation40
Results
EM of KaiC with a C-terminal bulky fusion can distinguish between the CI and CII rings
The crystallographic model of the KaiC hexamer with a resolution of ca. 3 Å reveals clear differences in the shapes of the N-terminal CI and C-terminal CII rings formed by domains that arise from a gene duplication (). Most prominent among the distinguishing features are the C-terminal peptides (residues 498–519) that protrude from the central region of the CII ring. By contrast, the C-terminal peptides in the CI half (residues 245–260) link the two rings at the waist and snake up on the outer surface of the hexameric particle. Further comparison of the shapes of surface models of the CI and CII hexameric rings makes evident the disk-like appearance of CI with more or less flat outer (N-terminal) and inner (turned toward CII) surfaces (). The CII ring is distinctly thicker and displays a strongly curved upper (C-terminal) surface with a dome-like appearance (). While these differences are readily apparent at the atomic resolution of the crystallographic model, they become blurred at resolutions of 20–30 Å in models derived from negative stain or cryo EM (e.g., refs. Citation15, Citation19, Citation25 and Citation28). Although it is tempting to associate flat and curved outer surfaces with the CI and CII end of the KaiC hexamer, respectively, the limited resolutions of the EM-based models render such distinctions unreliable. For example, class sums derived from electron micrographs of the binary KaiAC complex show the curved end of KaiC turned toward KaiA in some cases and the nearly flat end closer to KaiA in others. We know that KaiA protein is associated with the CII ring because of independent evidence that the dimer contacts a CII-terminal peptide.Citation14,Citation15 By contrast, the shapes of KaiC rings in EM class sums of KaiAC and KaiBC binary complexes do not provide sufficient evidence for making that distinction, thus giving rise to the uncertainty associated with the binding site of KaiB on KaiC based on low-resolution structural models alone.
We first used fusion proteins of S. elongatus KaiC and KaiB to establish that bulky additions allow one to distinguish between the CI and CII sides in KaiC electron micrographs. The KaiC protein featured the fluorescent Cerulean protein (MW ca. 26 kDa) at its C-terminal end and an additional C-terminal His6 tag. The KaiB protein was tagged with the fluorescent protein Venus (MW ca. 30 kDa). These constructs were previously used to quantify the rhythm of the KaiABC in vitro timer with FRET.Citation31 It is noteworthy that mixtures of these Kai fusion proteins with KaiA still generate regular rhythms of KaiC phosphorylation and dephosphorylation, indicating that the interactions among tagged proteins mimic those in the wild type oscillator. Negative stain EM micrographs of KaiC-Cerulean and of complexes between KaiC-Cerulean and KaiB-Venus indeed displayed buildup of density at one end. The KaiC ring adjacent to this additional density, therefore, had to be CII. However, flexibility of the fusion proteins relative to the KaiC barrel precluded class averaging and a visualization of the complexes in more detail.
Negative stain EM of KaiC-His6 labeled with 5 nm Ni-Nanogold
KaiC constructs with either N- or C-terminal His6 tags can be readily labeled with nickel (II) nitrilotriacetic acid gold nanoparticles (Ni-NTA-Nanogold).Citation32 Each Ni2+ is coordinated to an NTA and two histidines from the protein tag, thereby allowing attachment of the gold particle to specific sites on the protein surface and visualization by electron microscopy. Gold conjugation with the 5 nm probe was recently used in combination with an N-terminally His-tagged protein construct to discriminate between the N-terminal and C-terminal halves of the hexameric TrwK ATPase.Citation33 In our initial experiments, we used the 5 nm Ni-NTA Nanogold (Nanoprobes) with S. elongatus KaiC carrying either an N- or C-terminal His6 tag.
Negative stain electron micrographs of gold-labeled KaiC particles readily permit identification of the CI and CII rings. In the case of KaiC with the C-terminal His tag, the gold label is more or less centered above the CII ring (). This relative position is consistent with the orientations of C-terminal peptides relative to the KaiC double doughnut and shows that the tag is amenable to labeling for EM analysis (). Thus, C-terminal peptides protrude from the center of the dome, although their conformational flexibility allows for a range of gold positions above the CII dome. Variations in the position of the gold sphere relative to the CII ring could also be the result of complexation of the Ni2+ ion by His residues from two KaiC subunits. The Ni-NTA-Nanogold moiety appears to also fuse two or more KaiC hexamers in some cases. The variations in the relative positions of the gold sphere relative to KaiC particles coupled with the large size of the 5 nm gold particles made it difficult to generate interpretable class averages. These limitations prompted us to analyze the KaiBC complex with the smaller 1.8 nm Ni-NTA-Nanogold in combination with KaiC carrying a C-terminal His6 tag.
KaiB contacts the CII ring of KaiC
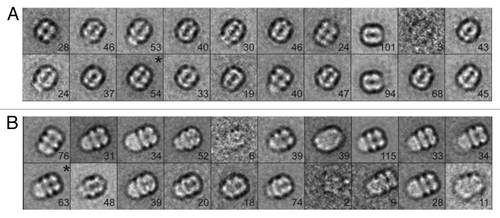
In order to locate the binding site of KaiB on KaiC, i.e., whether the KaiB dimer interacts with the CI ring or the CII ring, we bound the 1.8 nm Ni-NTA-Nanogold to the C-terminal His6 tag of S. elongatus KaiC S431D mutant. The S431D mutant (others have used T. elongatus KaiC S431E, e.g., ref. Citation22) is considered a mimic of the TpS431 KaiC phosphorylation state favored by KaiB, although the crystal structure of this mutant protein exhibited phosphorylation at T432 in three out of six subunits.Citation34 The tag and addition of the gold label are not expected to interfere with the function of KaiC, as interactions among Kai proteins involving the considerably more bulky Cerulean protein attached at the KaiC C-terminal end in combination with a His tag exhibited regular daily rhythms as assayed by FRET.Citation31 The small size of the 1.8 nm gold makes it difficult to see in the raw negative stain images; however, when negatively stained particles are averaged, the extra density of the gold can often be observed (see, for example ref. Citation35). As expected, negative stain electron micrographs of 1.8 nm gold-labeled KaiBC complexes do not show discrete gold spheres. However, in many cases there does appear to be extra diffuse density located on one side of the KaiBC complex (, arrows) that is not seen in images of the unlabeled particles (). It is possible that the binding of the Nanogold begins to restrict KaiC’s unstructured C-terminal tails, allowing these to be resolved by negative stain EM. Class sums were generated for both unlabeled and labeled KaiBC complexes. Class averages generated from unlabeled particles show the familiar three-layered structure with KaiB dimers forming the slightly thinner third layer (relative to the CI and CII rings) on one side of KaiC ( and ).Citation19,Citation28 Class sums of the gold-labeled KaiBC complex reveal a striking difference compared with reference particles in the form of a dense cloud located above the CII ring, undoubtedly representing the 1.8 nm gold particle(s) coated with KaiC His tags ( and ). We do not see discrete gold spheres in the averages due to the flexibility of the C-terminal tails and thus the position of the gold label. However, views approximately normal to the direction of the central KaiC channel show the characteristic cloverleaf-like appearance of KaiC, the KaiB third layer and a somewhat diffuse cloud generated by CII-terminal peptides, His tags and gold. These images leave no doubt that KaiB is bound on the same side as gold and, hence, contacts the CII ring. To separate the contributions of KaiB and gold tagging to the shape of the KaiBC complex in the negative stain EM class sums, we generated the difference density between gold-labeled KaiBC and gold-free KaiBC complex (). This analysis demonstrates convincingly that the difference density essentially corresponds to the C-terminal CII-His peptides in complex with Ni-NTA-Nanogold. The negative stain EM class sums also provide clear evidence that KaiB only binds at one end of KaiC, the CII side and not on both, as all images show co-location of KaiB and “gold cloud.”
Figure 4. Negative stain EM analysis of the S. elongatus KaiBC complex with and without 1.8 nm Ni-NTA-Nanogold. Representative micrographs of KaiBC particles either (A) unbound or (B) bound to 1.8 nm Ni-NTA-Nanogold. Arrows point to diffuse density not seen in images of unbound complexes. Scale bars equal 50 nm. Representative class sums of the complex between KaiB and KaiC-His6 in the (C) presence and (D) absence of 1.8 nm Ni-NTA-Nanogold. The numbers of particles included in each class is shown in the bottom right corner. (E) Difference mapping between averages depicted in panels (C and D), consistent with the presence of gold density adjacent to KaiB dimers. (F) The difference shown in E filtered at 3σ threshold. (C–F) Side length of panels, 240 Å.
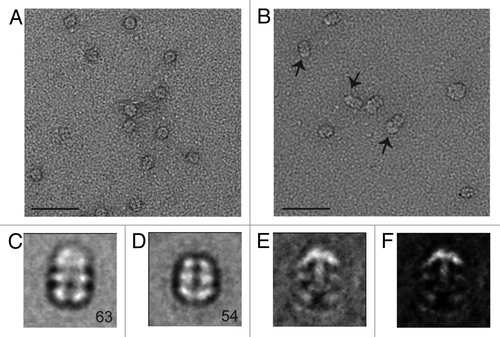
Figure 5. Analysis of S. elongatus KaiBC complex particles with and without 1.8 nm Ni-NTA-Nanogold. (A) Twenty class averages obtained by non-reference based alignment and classification of 875 unlabeled KaiBC particle images in negative stain. The average used for difference mapping (see ) is marked with an “*” in the upper right corner. (B) Twenty class averages obtained by non-reference based alignment and classification of 771 1.8 nm Ni-NTA-Nanogold labeled KaiBC particle images in negative stain. The average used for difference mapping (see ) is marked with an “*” in the upper right corner. (A and B) The number of particles in each projection average is shown in the lower right corner of each average. Side length of panels equals 240 Å.
Discussion
The goal of the present study was to remove lingering uncertainties as to the binding region of KaiB on the central clock protein KaiC. Our published model of the KaiBC complex based on negative stain and cryo EM placed KaiB at the CII end (see for CI and CII).Citation19 Although the resolution of the EM data was not sufficient to distinguish between the CI and CII rings, native PAGE assays revealed that mixtures of the separate CII hexamer and KaiB produced a bandshift, whereas mixtures of CI and KaiB did not. Although KaiB is a tetramer in solution (e.g., refs. Citation17 and Citation25), the protein binds to KaiC in the dimeric form, an observation first established by negative stain EM,Citation19 but confirmed by others.Citation21,Citation22 The central purpose of the additional work directed at the KaiBC complex and described here is not to determine a three-dimensional model (see refs. Citation19 and Citation25 for 3D-models of the KaiBC complex based on EM and SAXS, respectively), but rather to reject either CI or CII as the ring that is contacted by KaiB. Although one cannot a priori dismiss the possibility that KaiB can interact with both rings—perhaps at different time points during the 24-h clock cycle—different shapes and ESPs of the CI and CII rings () render this scenario rather unlikely. Indeed, the experimental data described here and employing gold-labeled KaiC protein are inconsistent with KaiB binding to both CI and CII, either simultaneously or at different times.
An important distinction between KaiC and many other AAA+ class proteins is that the hexamer in the cyanobacterial circadian clock is the result of a gene duplication,Citation6 whereby the N-terminal CI and C-terminal CII rings have discrete functions. Although it is useful to work with single-ring, hexameric proteins based on separately expressed recombinant CI and CII domains for biochemical and biophysical investigations, it is crucial to keep in mind that the physiological form of the KaiC protein in S. elongatus and T. elongatus is the dual-ring protein encompassing 519 and 518 amino acids, respectively. The KaiABC clock does not tick when either CI or CII are missing, and mixing separate CI and CII rings does not restore the rhythmicity seen with full-length KaiC in the presence of KaiA and KaiB. On the other hand, addition of steric bulk at the C-terminal ends of KaiC and KaiB does not interfere with the in vitro clock,Citation3 i.e., rhythmic phosphorylation and dephosphorylation of KaiC.Citation31 Thus, the KaiB-KaiC interaction is preserved even with a His6 tag and/or a 26 kDa protein (Cerulean) attached at the C-terminus of KaiC and/or a 30 kDa protein (Venus) attached at the C-terminus of KaiB.Citation31 This means that the KaiC construct used in the present work (C-terminal KaiC-His6 or KaiC-His6 S431D with or without gold tagging) can be expected to preserve the structural features of the wild-type KaiBC complex. Conversely, one cannot exclude the possibility that working with separate CI and CII hexamers or monomeric domainsCitation22 could result in non-specific interactions between the two proteins.
The negative stain EM data gathered for KaiBC complexes with gold ligated to the His tag at the C-terminus of KaiC provides unequivocal support for KaiB binding to the CII ring (). This new analysis confirms our previous models of the KaiBC complex derived from EMCitation19 and SAXSCitation25 that placed KaiB on the CII ring. Interestingly, the SAXS data considered alone are supportive of KaiB binding on the CII side, as the 3D envelopes constructed for KaiC hexamer alone and for the KaiBC complex revealed a protrusion on KaiC that is produced by CII-terminal KaiC peptides ( and ). In the SAXS model of the KaiBC complex, this protrusion is wedged between KaiB dimers (), consistent with the location of gold labeled C-terminal KaiC tails in the EM analysis presented here. KaiB dimers are therefore lodged in close proximity of the KaiA dimer that stimulates KaiC phosphorylation by binding to a CII-terminal peptide from a KaiC subunit ().Citation14,Citation15 There is no evidence that KaiB gets close to CI, and, instead, the KaiA protein becomes sequestered at the KaiB-KaiCII interface, preventing it from stimulating KaiC phosphorylation and bringing about dephosphorylation. Our previously published EM model of the ternary KaiABC complex derived in combination with native PAGECitation25 is consistent with the N-terminal KaiA domain bound to KaiBC at this stage, whereas C-terminal domains of KaiA dimer are involved in KaiC-peptide binding during the initial phosphorylation phase ().
Figure 6. Models of KaiC and KaiBC complex derived from small angle X-ray scattering (SAXS).Citation25 (A) SAXS model of S. elongatus KaiC hexamer, exhibiting a protrusion that marks the location of C-terminal peptides jutting out from the CII ring. (B) SAXS model of the KaiBC complex, consistent with KaiB dimers co-locating with C-terminal peptides and thus sitting on top of the CII ring.
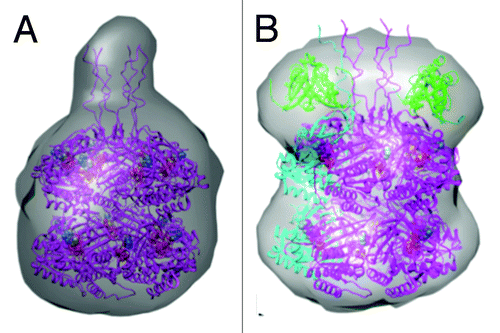
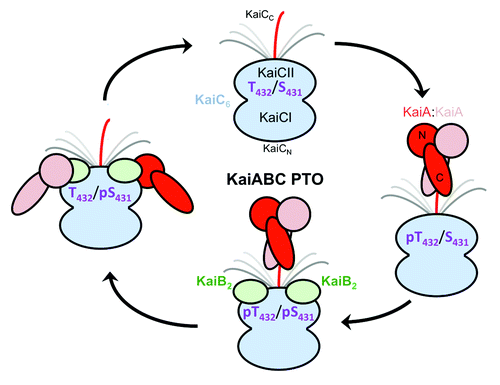
Figure 7. Changing protein-protein interactions over the daily cycle of the KaiABC PTO. KaiA dimer stimulates phosphorylation of KaiC in the hypo-phosphorylated T432/S431 (TS) state by interacting with a CII-terminal peptide via its C-terminal domains (C-KaiA). KaiC is first phosphorylated at T432 residues (pTS) and then also at S431 residues (pTpS = hyper-phosphorylated state). Unlike KaiA, KaiB shows no affinity for the KaiC C-terminal peptide and instead binds to the dome-shaped CII surface, triggering dephosphorylation first of pT432 (TpS) and then of pS431 residues via an ATP synthase mechanism, thus restoring the hypo-phosphorylated state. Dephosphorylation is accompanied by KaiC subunit exchange and the appearance of a ternary KaiABC complex that sequesters KaiA and therefore prevents it from stimulating KaiC phosphorylation. At this stage, KaiA lodges as a monomer on the side of the KaiBC complex, contacting it via its N-terminal (N-KaiA) domain. Each of the three proteins thus undergoes a change in quaternary structure over the 24-h period: KaiC hexamers exchange monomeric subunits, KaiB tetramer splits into dimers, and KaiA dimer splits into monomers.
We demonstrated earlier that SasA, a histidine kinase engaged in the clock output pathwayCitation36 and KaiB compete for similar binding sites on KaiC and derived a 3D-model of the SasA-KaiC complex from a hybrid structural approach (EM and SAXS).Citation25 Now that KaiB has been firmly established to bind to the CII ring of KaiC, it is evident that SasA also contacts that upper ring. Both proteins are sensing KaiC phosphorylation, KaiB in order to initiate subunit exchange and dephosphorylation and SasA to transmit the actual state of the oscillator to the clock-controlled gene expression pathways. The N-terminal sensory domain of SasA and KaiB exhibit similar folds,Citation37 although the former appears to contact CII as a monomer,Citation25 whereas the latter is a dimer. Similar functions (as far as sensing phosphorylation is concerned) and folds are therefore consistent with overlapping binding sites of the two proteins on KaiC. Because phosphorylation and dephosphorylation are limited to the CII half, it is reasonable to map the KaiB and SasA binding sites to CII.
The surfaces of the relatively flat CI ring (outer surface, , left) and the KaiCII dome (, right) exhibit starkly contrasting ESPs (almost uniformly negative, red, vs. mixed, red and blue, respectively). KaiB’s ESP does not feature highly positively polarized patches (), and the C-terminal end is very acidic (visible as a red hook in the panel on the far right). These observations also argue against KaiB (and therefore SasA) contacting the CI ring of an intact, full-length KaiC hexamer. Consistent with the importance of experiments using the physiologically relevant, complete form of KaiC to track protein-protein interactions, the evidence provided for SasA binding to CI is so far only based on the use of separate CI and CII domains.Citation29,Citation30 In summary, the work reported here, based on a refined EM analysis using gold-labeled KaiBC complexes, affirms that the KaiB protein binds to CII as do the KaiA and SasA proteins.
Materials and Methods
Protein expression and purification
S. elongatus KaiB and KaiC-His6 as well as the KaiC-His6 S431D mutant protein were expressed and purified following previously published procedures.Citation8,Citation19,Citation34 The KaiC C-terminal Cerulean-His6 and KaiB-Venus proteins were expressed and purified as described.Citation31
Preparation of KaiBC complex and incubation with gold
The S. elongatus KaiC S431D mutant (0.2 mg/mL) was incubated with S. elongatus KaiB (0.05 mg/mL) in the presence of AMPPnP (1 mM) overnight at 32°C. Dithiothreitol (DTT) was dialyzed from the KaiB-KaiC complex the next morning, and the sample was divided, with one half incubated with the Ni-NTA-Nanogold and one half used as a non-labeled control. The 1.8 nm and 5 nm Ni-NTA-Nanogold materials were purchased from Nanoprobes. Forty-three μmole of KaiBC complex were incubated with 65 nmole of the 1.8 nm Ni-NTA-Nanogold for 30 min at room temperature. The same procedure was used for incubating the complex with 5 nm Ni-NTA-Nanogold. The solution was diluted at the end of the incubation period. A volume of 1.2 μL of the sample was diluted with 8.8 μL of buffer (20 mM Tris, 100 mM NaCl, 1 mM ATP and 5 mM MgCl2). The final protein concentration used for EM grid preparation was 0.018 mg/mL.
EM grid preparation and electron microscopy
The carbon coated grids were glow discharged for better staining by decreasing the hydrophobicity of the grid. After applying the sample onto the grid, a wash buffer (10 mM imidazole, 20 mM Tris, 100 mM NaCl, 1 mM ATP and 5 mM MgCl2) was used to remove excess Nanogold per the protocol from Nanoprobes. Traces of buffer were then washed off with water. Uranyl formate (0.7% w/v) was used for conventional negative staining as described.Citation38 Images were taken under low-dose conditions at a magnification of 100,000× (defocus value of −1.5 mm) using a F20 electron microscope (FEI) equipped with a field emission gun at an acceleration voltage of 200 kV. Images were recorded on a 4 k × 4 k Gatan CCD camera, were converted to mrc format and binned by a factor of 2, resulting in final images with 2.0 Å/pixel. All binned MRC files were converted to SPIDER format using EM2EM.
Image processing and classification
Image analysis was performed with SPIDER and the associated display program WEB.Citation39 Particles of KaiB-KaiC either incubated or not incubated with Ni-NTA-Nanogold (Nanoprobes) were selected interactively and windowed into 120 × 120 pixel images (2.0 Å/pixel). Particles (875 unlabeled and 771 1.8 nm Ni-NTA-Nanogold labeled KaiBC particles) were rotationally and translationally aligned and subjected to 10 cycles of reference free alignment and K-means classification. Particles were grouped into 20 classes (). Difference images were calculated using both Diffmap.exe and SPIDER.Citation39 Results were similar using either program.
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM073845 (to M.E.). We are grateful to Prof Rama Ranganathan, Southwestern Medical Center, for providing us with the expression systems for the S. elongatus KaiC-Cerulean and KaiB-Venus fusion proteins, to Mr Said Sidiqi for help with protein expression and purification and to Mr Sabuj Pattanayek for assistance with ESP calculations.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol 2010; 8:e1000394; http://dx.doi.org/10.1371/journal.pbio.1000394; PMID: 20563306
- Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys 2011; 40:143 - 67; http://dx.doi.org/10.1146/annurev-biophys-042910-155317; PMID: 21332358
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 2005; 308:414 - 5; http://dx.doi.org/10.1126/science.1108451; PMID: 15831759
- Johnson CH, Egli M, Stewart PL. Structural insights into a circadian oscillator. Science 2008; 322:697 - 701; http://dx.doi.org/10.1126/science.1150451; PMID: 18974343
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 1998; 281:1519 - 23; http://dx.doi.org/10.1126/science.281.5382.1519; PMID: 9727980
- Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res 2000; 10:5 - 16; PMID: 10645945
- Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, et al. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA 2002; 99:17203 - 8; http://dx.doi.org/10.1073/pnas.262578499; PMID: 12477935
- Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell 2004; 15:375 - 88; http://dx.doi.org/10.1016/j.molcel.2004.07.013; PMID: 15304218
- Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA 2004; 101:13933 - 8; http://dx.doi.org/10.1073/pnas.0404768101; PMID: 15347809
- Egli M, Mori T, Pattanayek R, Xu Y, Qin X, Johnson CH. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry 2012; 51:1547 - 58; http://dx.doi.org/10.1021/bi201525n; PMID: 22304631
- Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem 2012; 287:18030 - 5; http://dx.doi.org/10.1074/jbc.M112.350660; PMID: 22493509
- Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J 2007; 26:4029 - 37; http://dx.doi.org/10.1038/sj.emboj.7601832; PMID: 17717528
- Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 2007; 318:809 - 12; http://dx.doi.org/10.1126/science.1148596; PMID: 17916691
- Vakonakis I, LiWang AC. Structure of the C-terminal domain of the clock protein KaiA in complex with a KaiC-derived peptide: implications for KaiC regulation. Proc Natl Acad Sci USA 2004; 101:10925 - 30; http://dx.doi.org/10.1073/pnas.0403037101; PMID: 15256595
- Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, et al. Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. EMBO J 2006; 25:2017 - 28; http://dx.doi.org/10.1038/sj.emboj.7601086; PMID: 16628225
- Egli M, Pattanayek R, Sheehan JH, Xu Y, Mori T, Smith JA, et al. Loop-loop interactions regulate KaiA-stimulated KaiC phosphorylation in the cyanobacterial KaiABC circadian clock. Biochemistry 2013; 52; In press PMID: 23351065
- Akiyama S, Nohara A, Ito K, Maéda Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol Cell 2008; 29:703 - 16; http://dx.doi.org/10.1016/j.molcel.2008.01.015; PMID: 18342562
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J 2003; 22:2127 - 34; http://dx.doi.org/10.1093/emboj/cdg212; PMID: 12505979
- Pattanayek R, Williams DR, Pattanayek S, Mori T, Johnson CH, Stewart PL, et al. Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation. EMBO J 2008; 27:1767 - 78; http://dx.doi.org/10.1038/emboj.2008.104; PMID: 18497745
- Hayashi F, Iwase R, Uzumaki T, Ishiura M. Hexamerization by the N-terminal domain and intersubunit phosphorylation by the C-terminal domain of cyanobacterial circadian clock protein KaiC. Biochem Biophys Res Commun 2006; 348:864 - 72; http://dx.doi.org/10.1016/j.bbrc.2006.07.143; PMID: 16901465
- Murakami R, Mutoh R, Iwase R, Furukawa Y, Imada K, Onai K, et al. The roles of the dimeric and tetrameric structures of the clock protein KaiB in the generation of circadian oscillations in cyanobacteria. J Biol Chem 2012; 287:29506 - 15; http://dx.doi.org/10.1074/jbc.M112.349092; PMID: 22722936
- Chang Y-G, Tseng R, Kuo N-W, LiWang A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc Natl Acad Sci USA 2012; 109:16847 - 51; http://dx.doi.org/10.1073/pnas.1211508109; PMID: 22967510
- Qin X, Byrne M, Mori T, Zou P, Williams DR, McHaourab H, et al. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc Natl Acad Sci USA 2010; 107:14805 - 10; http://dx.doi.org/10.1073/pnas.1002119107; PMID: 20679240
- Brettschneider C, Rose RJ, Hertel S, Axmann IM, Heck AJR, Kollmann M. A sequestration feedback determines dynamics and temperature entrainment of the KaiABC circadian clock. Mol Syst Biol 2010; 6:389; http://dx.doi.org/10.1038/msb.2010.44; PMID: 20631683
- Pattanayek R, Williams DR, Rossi G, Weigand S, Mori T, Johnson CH, et al. Combined SAXS/EM based models of the S. elongatus post-translational circadian oscillator and its interactions with the output His-kinase SasA. PLoS ONE 2011; 6:e23697; http://dx.doi.org/10.1371/journal.pone.0023697; PMID: 21887298
- Murayama Y, Mukaiyama A, Imai K, Onoue Y, Tsunoda A, Nohara A, et al. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J 2011; 30:68 - 78; http://dx.doi.org/10.1038/emboj.2010.298; PMID: 21113137
- Kageyama H, Nishiwaki T, Nakajima M, Iwasaki H, Oyama T, Kondo T. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell 2006; 23:161 - 71; http://dx.doi.org/10.1016/j.molcel.2006.05.039; PMID: 16857583
- Mori T, Williams DR, Byrne MO, Qin X, Egli M, Mchaourab HS, et al. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol 2007; 5:e93; http://dx.doi.org/10.1371/journal.pbio.0050093; PMID: 17388688
- Chang YG, Kuo NW, Tseng R, LiWang A. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA 2011; 108:14431 - 6; http://dx.doi.org/10.1073/pnas.1104221108; PMID: 21788479
- Valencia S J, Bitou K, Ishii K, Murakami R, Morishita M, Onai K, et al. Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells 2012; 17:398 - 419; http://dx.doi.org/10.1111/j.1365-2443.2012.01597.x; PMID: 22512339
- Ma L, Ranganathan R. Quantifying the rhythm of KaiB-C interaction for in vitro cyanobacterial circadian clock. PLoS ONE 2012; 7:e42581; http://dx.doi.org/10.1371/journal.pone.0042581; PMID: 22900029
- Reddy V, Lymar E, Hu M, Hainfeld JF. 5 nm Gold-Ni-NTA binds His tags. Microsc Microanal 2005; 11:Suppl 2 1118 - 1119; http://dx.doi.org/10.1017/S1431927605507712
- Peña A, Matilla I, Martín-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, et al. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem 2012; 287:39925 - 32; http://dx.doi.org/10.1074/jbc.M112.413849; PMID: 23035111
- Pattanayek R, Mori T, Xu Y, Pattanayek S, Johnson CH, Egli M. Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS ONE 2009; 4:e7529; http://dx.doi.org/10.1371/journal.pone.0007529; PMID: 19956664
- Passmore LA, Booth CR, Vénien-Bryan C, Ludtke SJ, Fioretto C, Johnson LN, et al. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol Cell 2005; 20:855 - 66; http://dx.doi.org/10.1016/j.molcel.2005.11.003; PMID: 16364911
- Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 2000; 101:223 - 33; http://dx.doi.org/10.1016/S0092-8674(00)80832-6; PMID: 10786837
- Vakonakis I, Klewer DA, Williams SB, Golden SS, LiWang AC. Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA. J Mol Biol 2004; 342:9 - 17; http://dx.doi.org/10.1016/j.jmb.2004.07.010; PMID: 15313603
- Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online 2004; 6:23 - 34; http://dx.doi.org/10.1251/bpo70; PMID: 15103397
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 1996; 116:190 - 9; http://dx.doi.org/10.1006/jsbi.1996.0030; PMID: 8742743
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 2001; 98:10037 - 41; http://dx.doi.org/10.1073/pnas.181342398; PMID: 11517324