Abstract
A key modality of non-surgical cancer management is DNA damaging therapy that causes DNA double-strand breaks that are preferentially toxic to rapidly dividing cancer cells. Double-strand break repair capacity is recognized as an important mechanism in drug resistance and is therefore a potential target for adjuvant chemotherapy. Additionally, spontaneous and environmentally induced DSBs are known to promote cancer, making DSB evaluation important as a tool in epidemiology, clinical evaluation and in the development of novel pharmaceuticals. Currently available assays to detect double-strand breaks are limited in throughput and specificity and offer minimal information concerning the kinetics of repair. Here, we present the CometChip, a 96-well platform that enables assessment of double-strand break levels and repair capacity of multiple cell types and conditions in parallel and integrates with standard high-throughput screening and analysis technologies. We demonstrate the ability to detect multiple genetic deficiencies in double-strand break repair and evaluate a set of clinically relevant chemical inhibitors of one of the major double-strand break repair pathways, non-homologous end-joining. While other high-throughput repair assays measure residual damage or indirect markers of damage, the CometChip detects physical double-strand breaks, providing direct measurement of damage induction and repair capacity, which may be useful in developing and implementing treatment strategies with reduced side effects.
Introduction
Ionizing radiation (IR) and genotoxic chemotherapeutics are frontline tools in cancer management.Citation1,Citation2 One of their main mechanisms of action is the formation of toxic double-strand breaks (DSBs) that can inhibit cell division and induce cell death in tumor cells. Normal mammalian cells rely predominantly upon two major pathways of DSB repair: non-homologous end-joining (NHEJ) and homologous recombination (HR).Citation3-Citation5 These repair pathways reduce the toxicity of these treatments and are also known to modulate sensitivity of tumors to chemotherapeutics. For example, DSB repair has been identified as an underlying mechanism of drug resistance and is also important in guiding treatment strategies that more selectively target cancerous cells and reduce side effects.Citation6,Citation7 Ironically, although we use DSB inducing agents to treat cancer, we also know that spontaneous and environmentally induced DSBs are an important risk factor for cancer susceptibility. Thus, the ability to evaluate DSBs is relevant both for cancer treatment and cancer prevention. An emerging approach for treating cancer is to sensitize tumors by inhibiting their DNA repair response system, e.g., NHEJ.Citation8-Citation11 A major challenge in identifying such inhibitors is that currently available DNA damage assays are limited in throughput and often provide information about residual damage (i.e., chromosomal aberrations) but offer little insight into the actual lesion burden or kinetics of repair. Better methods to directly measure DSBs could therefore be useful for assessing a person’s DNA repair capacity (relevant to cancer susceptibility), assessing DNA repair capacity in tumor cells (so as to predict drug sensitivity) and for identifying novel pharmaceutical compounds.
Currently, one of the most broadly used approaches for assessing DSBs is to measure the levels of phosphorylated serine 129 of the histone variant H2AX (γ-H2AX), an early signaling event in response to a DSB. Although the γ-H2AX assay is remarkably sensitive,Citation12 H2AX phosphorylation is separable from DSBs, in part due to its dependence on the activity of ATM, DNA-PK and other phosphatidylinositol 3-kinase (PI3K)-related kinases (PI3KKs).Citation13 An alternative approach is to directly measure DSBs based on their physical properties. Direct physical detection of DSBs prevents problems that are associated with quantifying cellular responses and is thus considered to be the gold standard.
Physical detection is the basis for both the alkaline elution method and the neutral single-cell gel electrophoresis assay (known as the neutral comet assay), both of which rely upon changes in the mobility of intact vs. broken DNA.Citation14,Citation15 Each of these approaches has serious limitations, however. The alkaline elution method suffers from being technically difficult and slow, and thus is used increasingly rarely. Although there are many reports of the neutral comet assay being used for analysis of DSBs,Citation16-Citation18 unlike its alkaline counterpart, which is well accepted for analysis of single-strand lesions, the neutral comet assay is a highly controversial approach. Some argue that the approach does not provide the resolution required for detailed DSB analysis.Citation19,Citation20 Equally problematic is the issue of throughput and noise. The traditional neutral comet assay suffers from very low throughput, and high sample-to-sample variation (estimated to be as high as 26% inter-scorer and 47% inter-laboratory).Citation21,Citation22 If these limitations were overcome, the neutral comet assay could enjoy the same success as the alkaline version for detection of base damage and single-strand damage.
With an interest in leveraging the comet assay for broader applications, we recently developed the CometChip platform, which exploits microfabrication to enable the creation of a single-cell microarray. The arrayed cell format () has been shown to help overcome problems in reproducibility and throughput of the comet assay;Citation23 however, the platform had not previously been shown to be effective for analysis of DSBs. Here, we set out to: (1) evaluate the specificity of the neutral CometChip for detection of DSBs, (2) demonstrate efficacy of a 96-well plate format and (3) evaluate the potential advantages of the multi-cell/microwell format both in terms of throughput and sensitivity. Using a combination of cell lines carrying specific DSB repair defects, as well as chemical inhibitors of DSB repair, we show that the neutral CometChip can indeed be used as a tool for monitoring DSB formation and repair. In addition, we show here that throughput is increased by using a 96-well plate format (compatible with high-throughput screening tools) and by exploiting multi-cell comets (enabled by capturing multiple cells within a single microwell), which dramatically improve both throughput and sensitivity. Indeed, experimental conditions that traditionally require ~100 individual images, each analyzed separately, can now be performed with just a few images (without loss of sensitivity). Together, these advances make it possible for the first time to physically monitor DSB formation and repair in a high-throughput fashion, thus enabling evaluation of DSB formation in the clinic, in the pharmaceutical industry and in epidemiological studies.
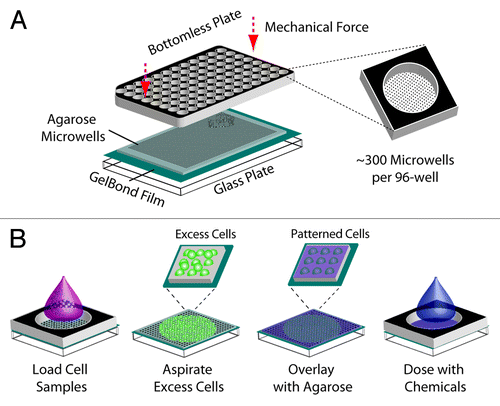
Figure 1. Creation of a 96-well macrowell array. (A) Assembly of macrowell comet array. Agarose gel with microwells is sandwiched between a glass substrate and a bottomless 96-well plate and sealed with mechanical force. Approximately 300 arrayed microwells comprise the bottom of each macrowell. (B) Loading and chemical dosing of cell samples in macrowell. One sample is loaded into each macrowell and cells settle by gravity into the arrayed microwells. The bottomless plate is removed in order to aspirate excess cells and enclose the cells in agarose. The bottomless plate is replaced in order to treat each macrowell with a chemical condition.
Results
High-throughput neutral comet assay
Currently available assays for detecting DSBs are limited in throughput and speed of analysis, attributes that are necessary for development of effective clinical or pharmaceutical assays and for epidemiological studies. We recently developed the CometChip, which enables up to 96 different cell samples or chemical conditions to be analyzed on a single agarose gel (). To perform the assay, cells are arrayed in microwells in an agarose gel by gravity and subsequently cultured, treated, lysed and electrophoresed using traditional comet protocols.Citation15,Citation24 The resulting patterned comets at the base of each well of the 96-well plate, hereafter referred to as a macrowell (), can be imaged manually on a fluorescent microscope or in an automated fashion using an imaging platform. One advantage of the arrayed microwell approach is that the comets are optimally patterned at the same focal depth with no overlap, enabling 10 or more comets to be captured and automatically analyzed in a single image, rather than one at a time using the traditional method. Images can also be analyzed using unbiased, fully automated software, reducing the processing time from hours to minutes.
Figure 2. Arrayed microwell comet assay for detection of double-strand breaks. (A) Arrayed microwell comets from untreated TK6 human lymphoblasts and TK6 cells exposed to 100 Gy IR. Scale bar is 100 μm. (B) Comparison of irradiation dose response between traditional comet slides scored using commercial software and microwell comets scored using automated software. Each data point is the average of three independent experiments, where the median percent total DNA in tail (top), the median olive tail moment (middle), or the median tail length (μm) of 100 individual comets were used to represent the extent of DNA damage. Error bars represent the standard error of the mean from the four independent experiments. % DNA tail, R2 = 0.92 (CometChip) and R2 = 0.42 (traditional); olive tail moment, R2 = 0.97 (CometChip) and R2 = 0.63 (traditional); tail length (μm), R2 = 0.94 (CometChip) and R2 = 0.85 (traditional). (C) Bleomycin dose response conducted on macrowell platform with each data point representing the median tail length (μm) of at least 300 comets pooled from six macrowells.
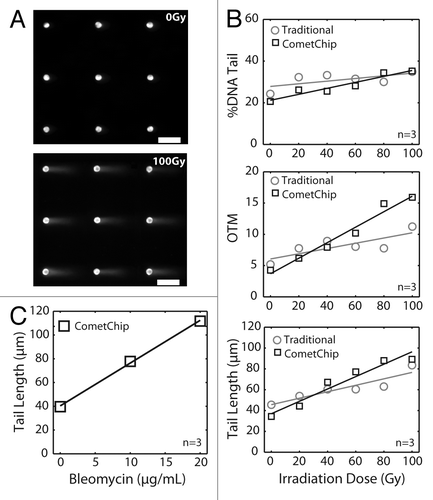
To explore the potential utility of the CometChip for studies of DSBs, we compared the traditional neutral comet analysis to the CometChip in cells exposed to ionizing radiation (IR). DNA damage can readily be observed by the change in morphology from a sphere (undamaged nucleus, , top) to a comet-like morphology (, bottom). Standard methods for assessing DNA damage from measurement of DNA signal in comet tails include comet tail length (μm), percent tail DNA and comet tail moment. A comparison of the traditional assay and the CometChip platform shows that both approaches yield a linear dose response across the 0–100 Gy range (). Among the three standard comet measurement parameters, it was found that tail length is the most sensitive parameter (R2 > 0.8 for both traditional assay and the CometChip). As expected, the absolute values vary somewhat between the traditional and the CometChip approach. Importantly, however, the CometChip is consistently more sensitive, as reflected by the regression statistics (for tail length, R2 = 0.85 and R2 = 0.94 for the traditional and CometChip data, respectively). This increased sensitivity may result from the improved morphology of the microwell comet, since the DNA is constrained by the walls of the microwells, providing a more distinct head-to-tail cutoff and, thus, more reliable assessment of comet tail length. It is noteworthy that for the traditional approach, each data point in corresponds to comets collected from several glass slides. Consequently, a total of 30 slides were analyzed for the traditional assay vs. a single CometChip.
To further assess the efficacy of the neutral CometChip for evaluating chemical exposures, we assayed bleomycin, a glycopeptide antibiotic that efficiently introduces DSBs and is used in the treatment of numerous cancers.Citation25–Citation27 To analyze bleomycin-induced DSBs, more than 300 comets were pooled from six replicate macrowells for each data point, and the resulting data fit a linear dose response (). Together, these data show that the CometChip may be used to measure DSBs following multiple chemical exposure conditions in parallel.
Statistical analysis and reproducibility
Traditionally, the comet assay is performed by putting a layer of agarose onto a glass slide. This approach introduces noise from several sources, including variation in focal plane, overlapping comets, non-uniformity of electrophoresis conditions. By placing cells in a single focal plane, preventing overlapping comets via the microarray and performing analysis of 96 samples in parallel, experimental noise is greatly reduced.
Previously, we had shown that by increasing the size of the microwell, it is possible to capture multiple cells per microwell and furthermore that the cluster of cells can be effectively analyzed as a single “microwell-comet.”Citation23 The microwells essentially provide uniform comet head, making the system self-calibrating and thus enabling analysis of multi-cell microwells, each as a single comet. To learn more about how data from multi-cell microwell comets compares to traditional analysis, we plated human lymphoblastoid cells into microwells of various sizes (from 25 μm, which generally captures a single cell, to 45 μm, which captures multiple cells per well) (), and we analyzed both the sensitivity and the comet-to-comet variation following exposure to a range of doses of IR. Analysis of tail length shows that the sensitivity is not significantly affected by the size of the microwell (). This result is consistent with earlier studies under alkaline conditions.Citation23
Figure 3. Statistical analysis and reproducibility of the CometChip compared with traditional comet slides. (A) IR dose response of TK6 cells loaded into different sized microwells. Each data point is the average of three independent experiments, where the median tail length of at least 100 individual comets was obtained in each experiment. Fluorescent images display 25, 30 and 45 μm diameter microwells filled with Syber Gold stained TK6 human lymphoblasts. Scale bar is 10 μm. (B) TK6 cells were irradiated with 100 Gy IR. Randomly selected 10, 50 or 100 individual comets on traditional glass slides or microwell comets on the CometChip were analyzed. Each data point is a single comet or microwell comet. Box plots show median tail length (μm) of the data set as the middle line and the lower and upper quartiles as the box. Whiskers show extent of furthest data points within 150% of interquartile range. (C) Slide-to-slide variability of traditional comet assay and macrowell-to-macrowell variability of 96-well format CometChip using TK6 cells exposed to 100 Gy IR. For traditional slides, medians of 30 traditional neutral comets from 10 slides are plotted. Mean comet length of 10 sides, 69 μm; coefficient of variation, ~20%. For CometChip, medians of at least 50 microwell comets from 10 macrowells of the CometChip are plotted. Mean comet length of 10 macrowells, 109 μm; coefficient of variation, ~5%.
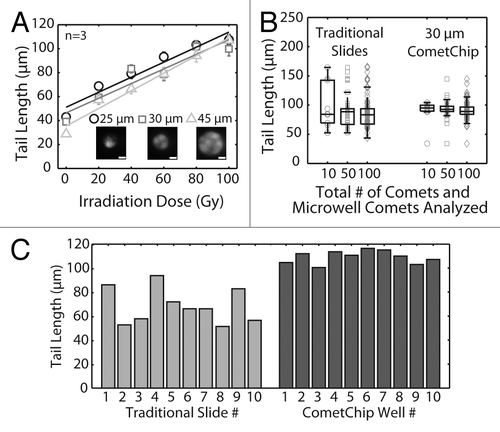
Given that multiple cells contribute to a single data point (which essentially averages the data for multiple cells in a single multi-cell comet), we predicted that the variance among multi-cell comets would be lower than that of traditional comets. To explore this possibility, we measured the tail length for individual cells and for individual multi-cell microwell comets. Among individual traditional comets (which are, by definition, single cells), tail length is highly variable, ranging from about 50 to about 175 um. For the traditional comets, as the sample size increases from 10 to 100 comets, the range between the upper and lower quartiles is reduced, as expected. Remarkably, analysis of only 10 microwell comets reveals data that is tightly clustered, with upper and lower quartile ranges that are even smaller than the data from 100 traditional comets (; median is indicated by the middle line, while the box indicates the upper and lower quartiles). Likewise, analysis of standard deviation (data now shown) again indicates that analysis of just 10 microwell comets yields data with a smaller standard deviation than that of 100 traditional comets. Given that nine multi-cell comets can be captured in a single image (), it is now possible to collect data that is equivalent or better than that from 100 traditional images, in just two images.
Having observed that the CometChip platform greatly reduces comet-to-comet variance, we next asked if the platform is also advantageous with regard to sample-to-sample variation. Here we compared samples from 10 macrowells of the 96-well plate of the CometChip to 10 traditional slides. TK6 lymphoblastoid cells were exposed to 100 Gy ionizing radiation (IR) and analyzed for DSBs using the two platforms. As expected, traditional assay performed on glass slides demonstrated high variability from slide to slide (), with a coefficient of variation of around 20% (lower than the published estimates, because only one scorer performed the experiment here). Conversely, 10 macrowells of the CometChip displayed a coefficient of variation of only 5% (), which is not increased even when we extend the analysis to the entire chip (Fig. S1). Taken together, being able to process 96 samples in parallel on a single platform, rather than on separate slides, is not only more practical, but it also greatly suppresses experimental noise.
Detection of DSBs on the CometChip
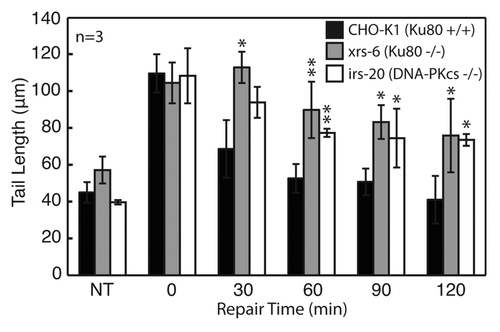
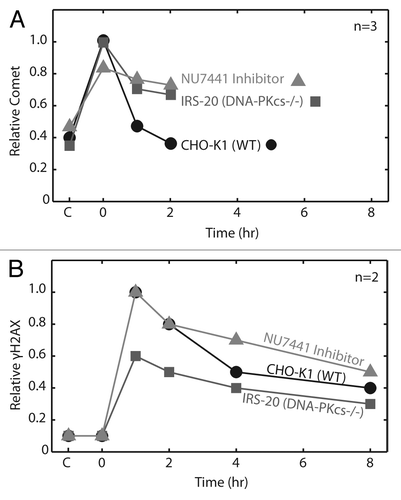
In order to assess the ability of the neutral comet to detect DSBs, we set out to compare DSB repair kinetics among cell lines with known genetic deficiencies in NHEJ. An early step in the NHEJ pathway is recognition of the DSB by the Ku70/Ku80 heterodimer. The Ku heterodimer recruits the DNA-PK catalytic subunit, DNA-PKcs, to form the DNA-PK complex, which promotes strand alignment and recruitment of factors involved in end-processing and ligation.Citation28,Citation29 We used mammalian cells specifically mutated in either Ku80 (xrs-6) or DNA-PKcs (irs-20) to see if cells lacking critical genes in NHEJ show a repair deficit when analyzed on the CometChip. Cells were exposed to 100 Gy IR, and the macrowells corresponding to the zero min time point were immediately lysed, while the remaining macrowells were filled with growth medium and allowed to repair over the course of 120 min. Wild type cells demonstrated a fast recovery within the first hour, while both the Ku80- and DNA-PKcs-deficient cells showed a severe repair defect, with about half of the damage still remaining after 2 h (). These data show that the assay is effective for detection of DSBs, which is consistent with previously published studies of Ku80 and DNA-PKcs deficient cells using similar methods.Citation30,Citation31 Interestingly, even in the absence of DNA-PK activity, there is some residual clearance of DNA damage, which is likely due to alternative DSB repair pathways that are revealed in the absence of NHEJ.Citation3,Citation32
Figure 4. Evaluation of DNA repair kinetics of CHO-K1 (wild type), xrs6 (Ku80−/−) and irs20 (DNA-PKcs−/−) exposed to 100 Gy IR. All cell types and repair times conducted on a single CometChip with data representing median comet tail lengths (μm) from at least 50 comets. Error bars represent standard deviations of three independent experiments. Symbols indicate a significant difference compared with wild type according to t-test: *p < 0.05, **p < 0.005
Comparison of CometChip to the γ-H2AX assay
One potential advantage of the CometChip is that it enables direct physical measurement of DSBs, rather than assaying for an indirect marker of a double-strand break, such as γ-H2AX. To directly compare CometChip and the γ-H2AX assay, we studied repair following exposure to 100 Gy IR. Physical breaks were assessed by CometChip and levels of γ-H2AX were determined by western blot, as shown in , respectively. Exposure to IR leads to almost immediate creation of DSBs, and these breaks are detectable with the CometChip. In contrast, DSBs are not detected using γ-H2AX assay immediately after exposure, which is consistent with literature showing that it takes time for foci to form (γ-H2AX signal peaks and plateaus 10–30 min post exposure).Citation33 Furthermore, whereas there is rapid clearance of DSBs within about 2 h of exposure, as shown by the ComeChip analysis, the apparent rate of clearance is much slower for γ-H2AX. Indeed, the signal for DSBs persists for several hours after DSBs have been completely repaired according to the CometChip analysis. Thus, the appearance and the disappearance of the γ-H2AX signal clearly lags behind repair of actual physical DSBs, and interpretations of results from this assay based on single time points could be misleading, because the measurements do not directly reflect the actual kinetics of DSB repair. Taken together, these data demonstrate the value of physical detection of DSBs rather than assessment of a response to the damage. Furthermore, they support the use of the CometChip for analysis of small-molecule inhibitors of DNA-PK and other DSB repair proteins.
Figure 5. Comparison of neutral CometChip to γ-H2AX assay for repair kinetics. Wild type cells were exposed to 100 Gy IR. (A) All cell conditions and repair time points were conducted in triplicate macrowells of a single CometChip. Comet tail length (μm) values were normalized to peak wild type damage. (B) Repair kinetics were measured in triplicate over eight hours using a western blot version of the γ-H2AX assay. Error bars represent standard deviation of three replicate experiments.
Screen of DNA-PK inhibitors
Evaluating DNA-PK inhibitors that could be useful in the clinic is a potentially valuable application of CometChip technology, as inhibitors of DSB repair are of particular clinical values as radio- and chemotherapy sensitizers. To test the efficacy of the CometChip as a tool for identifying repair inhibitors, we screened a subset of leading DNA-PK inhibitors, listed in . Wortmannin and LY294002 are benchmark PI3KK inhibitors that have been shown to sensitize cancer cells to DNA damaging agents via inhibition of DNA-PK-dependent DSB repair.Citation34-Citation36 NU7441, NU7026 and Compound 401 were developed as LY294002-like inhibitors with improved specificity for DNA-PK.Citation37-Citation39 DMNB and PI103 reflect the range of specificity and efficiency of other DNA-PK inhibitors, with DMNB having high selectivity and relatively low inhibitory efficiency for DNA-PK (i.e., 15,000 nM), and PI103 having nanomolar potency for numerous PIKKs.
Table 1. DNA-PK inhibitors
CHO-K1 (wild type) cells were loaded into a 96-well CometChip and exposed in triplicate macrowells to 50 µM of each inhibitor. This concentration represents the half maximal effective concentration (EC50) of LY294002, which can be greater than 1,000-fold higher than the half maximal inhibitory concentration (IC50), due to cellular penetration, competition with ATP and the high abundance of cellular DNA-PK.Citation40 The use of the 96-well plate minimized the volume requirement of each inhibitor to 50 µL and reduced the complexity of the experiment to just three CometChips. All steps including cell loading, repair inhibition, irradiation and repair took less than three hours to complete. Imaging was greatly reduced in terms of labor requirement through automation.
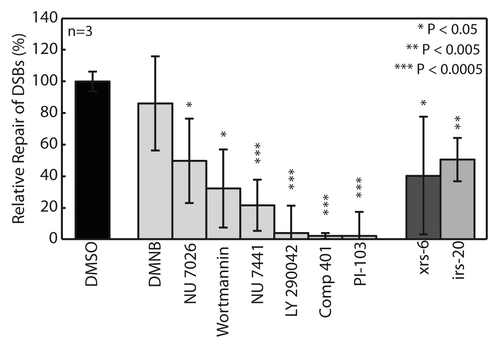
NU7441 is being pursued as a potential adjuvant in cancer treatment due to its ability to increase efficacy of both chemo- and radiotherapy in tumor-bearing mice through selective inhibition of DNA-PK.Citation41 shows the relative repair 1 h after exposure compared with wild type for each inhibitor and cell type. All inhibitors, except for the less potent DMNB, displayed inhibition of DSB repair at an efficiency comparable to or greater than the cells with DNA-PK genetic deficiencies. One possible reason for reduced repair relative to NHEJ-deficient cells is that some inhibitors have multiple PI3KK targets, which could lead to a more pronounced effect on DSB repair. Inhibition of mammalian target of rapamycin (mTOR) has recently been shown to significantly inhibit NHEJ, which might explain the increased efficacy of several of the DNA-PK inhibitors.Citation42 Most notably, Compound 401 targets DNA-PK and mTOR, and was found to completely inhibit DSB repair. Thus, the CometChip can be used to both identify inhibitors of DSB and provide information regarding their relative potencies.
Figure 6. Evaluation of relative repair from 100 Gy IR after 1 h exposure to DNA-PK inhibitor library. CHO-K1 (wild type) cells pre-incubated for 1hr in 50 μM of each inhibitor. All conditions and controls, xrs6 (Ku80−/−) and irs20 (DNA-PKcs−/−), assayed in triplicate macrowells. Data and error bars represent averages and standard deviations of three independent experiments. Symbols indicate significance compared with wild type (DMSO) according to t-test: *p < 0.05, **p < 0.005, ***p < 0.0005.
Discussion
Here, we have described a high-throughput method to directly assess DSBs resulting from both radiation and a chemotherapeutic agent, and we show the DSB repair kinetics for multiple cell samples analyzed in parallel. DNA-damaging agents are central to non-surgical cancer treatment and as adjuvants to surgery, but their clinical efficacy varies considerably among individuals. The CometChip provides a relatively inexpensive and fast method of measuring DSB repair capacity, which could be used to assess patient sensitivity and tumor resistance in order to design optimal therapeutic strategies that minimize side effects. Additionally, an assay for DSBs in human cells can be used to assess environmental exposures and interindividual variation in susceptibility to exposures that induce DSBs.
The morphology of neutral comets differs from that of alkaline comets and is often criticized for reducing the sensitivity and reproducibility of the assay. Single-strand breaks relax supercoiled DNA, resulting in a “halo” that makes it difficult to decipher the head-to-tail threshold.Citation15 One advantage of the arrayed comet platform is that the DNA conforms to the morphology of the microwell, producing a distinct and reproducible comet head and tail. This feature may explain the increased sensitivity of the CometChip compared with the traditional comet assay as determined by ANOVA analysis. Another contributing factor may be the reduced variability in the CometChip. A major advantage of the CometChip is the reduction of labor, and therefore human error, resulting from the ability to conduct all samples and controls on a single device integrated with high-throughput screening technologies. With this system, we are now able to investigate smaller differences among samples, which were once overwhelmed by slide-to-slide variations in the traditional assay. Also, because the CometChip is fully integrated with automated imaging and analysis platforms, high inter-scorer and inter-laboratory variability observed in traditional assay would be significantly reduced.Citation21,Citation22 In addition to increased sensitivity and reproducibility, it is noteworthy that the CometChip, by allowing multi-cell comets analysis, enables assessment of DNA damage in cell aggregates, which more realistically reflect biological systems.
High-throughput screens are most useful when a very small volume is required per sample, thus minimizing the amount of test agent required. An important feature of the CometChip is that the demand for chemical reagents is significantly reduced compared with the traditional assay (while ~2 ml is generally required for on-slide treatment using the traditional method, only ~50 μl is required using the CometChip per treatment condition). This is important in terms of cost savings when evaluating expensive chemotherapeutic compounds. Furthermore, all chemical conditions can be conducted on the CometChip, eliminating the need for cell plating and post-exposure centrifugation and trypsinization required by other high-throughput versions of the comet assay (Trevigen, Inc.).Citation43 The increase in throughput not only has potential applications in drug screening and personalized medicine, but it also can be used to better classify environmental pollutants and understand the risk they represent to exposed populations.
In addition to NHEJ and HR, DSBs can also be repaired by alternative mechanisms, such as single-strand annealing (SSA) and microhomology mediated endjoining (MMEJ).Citation4,Citation32 In the case of HR, we do not anticipate detecting a significant impact from this pathway during the early time points, since NHEJ is significantly faster than HR.Citation3 Likewise, SSA and MMEJ are significantly slower than NHEJ and are thus kinetically separable. Indeed, we observed residual end joining with slower kinetics in the NHEJ mutant cells (), which is consistent with residual repair by SSA, MMEJ and possibly HR. The ability of the CometChip to measure rapid repair during the initial 2-h repair period provides a tool for studying factors that impact classical NHEJ.Citation32
The irs-20 CHO cell line contains a defect in the DNA-PKcs that disrupts its kinase domain, while maintaining its DNA-binding activity.Citation44 Detection of this deficiency supports the use of the CometChip for screening potential adjuvant therapies known to inhibit the phosphorylation activity of DNA-PK. Indeed, we found that the potent DNA-PK inhibitor NU7441 induces equivalent levels of repair inhibition as the irs-20 mutant (). Interestingly, compounds with lower specificity for DNA-PK demonstrated stronger inhibition profiles, suggesting the potential importance of other PI3KKs in DSB repair mediation. This is best exemplified by LY290042, which has relatively low potency for DNA-PKcs compared with the other inhibitors (), but had a dramatic effect on DSB repair capacity. In addition to potential differences due to non-specific targets, the increased efficacy of LY290042 over wortmannin may also result from its ability to competitively inhibit the active site of DNA-PKcs, as opposed to the non-competitive nature of wortmannin.Citation45 Further differences in efficacy may be due to off-target interactions with mTOR, as previously discussed,Citation42 or PI3K, which has recently been shown to influence DSB repair through interaction with other PI3KKs (i.e., ataxia telangiectasia mutated).Citation45 In support of this hypothesis, NU7026, a highly selective inhibitor, has significantly less potency for mTOR and negligible affinity for PI3K and did not suppress repair as effectively as NU7441 (). It will be interesting to further explore this concept through evaluation of specific inhibitors of mTOR, such as PP242 and PP30.Citation42 There are limited tools to investigate the direct effect of the PI3KK signaling cascade and other chromatin remodeling processes on DSB repair. Chromatin modifications are gaining attention for their role in carcinogenesisCitation46 and for their potential as targets for pharmaceutical intervention.Citation48 CometChip studies are currently underway to assess other PI3KK-targeting compounds, such as ATM (i.e., KU55933). Additional studies are also underway to investigate histone deacetylase (HDAC) inhibitors, a promising new class of small molecules in cancer treatment.Citation49
In conclusion, the CometChip is an effective tool for directly measuring the induction of DSBs. The 96-well format enables multiple cell types and chemical conditions to be assayed in parallel with improved processing speed to that of the traditional comet assay. Using both a genetic approach and a chemical inhibitor of DNA repair, we demonstrated that the assay is sensitive for DSB detection and showed that the assay improves reproducibility and enhances throughput by nearly an order of magnitude compared with the traditional assay according to reference Citation15. Furthermore, the platform directly measures DSB repair kinetics, enabling the detection of end-joining deficiencies and the evaluation of inhibitors of DSB repair. It is noteworthy that this new DNA damage-sensing platform opens doors to studies that were previously virtually impossible. The ability to screen 96 samples in parallel with significantly reduced noise enables detection of very subtle differences among cell types and exposures. Additionally, the reduced processing time from months to days potentiates large-scale screening. Taken together, it is hoped that the neutral CometChip will give rise to new insights into the roles of DNA damage and repair in health. (AUTHOR: Please cite ref. Citation47 in main text)
Materials and Methods
Cell culture
TK6 human lymphoblastoids were cultured in suspension in 1× RPMI medium 1640 with L-glutamine (Invitrogen) supplemented with 10% horse serum (Invitrogen). Chinese hamster ovary (CHO) cell lines were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals). All cell culture media were supplemented with 100 units/mL penicillin-streptomycin (Invitrogen). CHO-K1 and xrs-6 CHO cells were kindly supplied by Larry H. Thompson (Lawrence Livermore National Laboratory), and irs-20 CHO cells were a generous gift by Dr. Joel S. Bedford (Colorado State University). All genotypes were confirmed by western blot (data not shown).
Microwell fabrication
Negative molds were fabricated from silicon wafers (WaferNet) that were lithographically patterned with SU-8 photoresist microwells (SU-8 2025, Microchem), following a protocol similar to Wood, et al.Citation15 Polydimethylsiloxane (PDMS, Dow Corning) was cast on the negative silicon mold and baked for 1 h at 50°C according to the manufacturer’s instructions. After 1 h, the PDMS was removed to reveal patterned microposts of approximately 50 μm depth and 30 μm width. Molten 1% normal melting point agarose (Omnipur, Invitrogen) was applied to a sheet of GelBond film (Lonza), and the PDMS mold was allowed to float until the agarose set. The PDMS mold was removed, leaving a 300 μm thick gel with arrayed microwells. The microwell gel was then clamped between a glass plate and a bottomless 96-well titer plate (Greiner BioOne).
Macrowell preparation
illustrates CometChip loading. At least 10,000 cells were added to each macrowell (well in a 96-well plate) and allowed to settle by gravity in complete growth media at 37°C, 5% CO2. Excess cells were aspirated after 15 min, and the bottomless 96-well plate was removed in order to enclose the arrayed cells in a layer of 1% low melting point agarose. Traditional comet slides were prepared as previously described.Citation15
Exposure to ionizing radiation
Microwell gels were either irradiated all at once in the 96-well format, or cut and exposed in smaller pieces. A Cobalt-60 irradiator (Gammacell 220 Excel, MDS Nordion) was used to deliver 0–100 Gy of ionizing radiation (IR) at an approximate rate of 120 Gy/min. Cells were irradiated in 4°C phosphate buffered saline.
Treatment with chemicals
Bleomycin (Bioworld) was dissolved in phosphate buffered saline and was exposed to cells for 1 h at 4°C. All chemical conditions, controls and replicates were conducted on a single CometChip. All repair inhibitors were purchased from Tocris. NU7441, NU7026 and PI103 hydrochloride were prepared in dimethyl sulfoxide (DMSO) and Wortmannin, LY294002, Compound 401 and DMNB were prepared in ethanol. All inhibitors were stored at -20°C until immediately before use. The inhibitors were diluted to 50 μM in complete growth medium and exposed in triplicate macrowells for 1 h at 37°C, 5% CO2. CHO-K1 cells in 1% DMSO and untreated xrs-6 and irs-20 cells were used as controls. The inhibitors were replaced with 4°C phosphate buffered saline during irradiation and fresh inhibitor was applied during evaluation of repair kinetics.
Repair on-chip
To evaluate repair kinetics, cells were treated and then incubated in media to permit repair. At various times after initiation of repair, repair was stopped by lysis (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris, 1% N-Lauroylsarcosine, pH 9.5 with 0.5% Triton X-100 and 10% DMSO added 20 min before use). The 0 min repair time point was achieved by adding lysis buffer to macrowells immediately post exposure. Multiple repair times were analyzed on a single plate by adding lysis buffer to the macrowells at the appropriate times. After all cells have been lysed, the entire gel was removed from 37°C incubator, immersed in fresh lysis buffer and transferred to a 43°C incubator. In the evaluation of NHEJ deficient cells, all time points (0, 30, 60, 90, 120 min) were conducted on a single CometChip (with 96 macrowells formed using a bottomless 96-well plate). For the screen of NHEJ repair inhibitors, each condition (nontreated, 0 and 60 min) was run on a separate gel.
Neutral comet assay
The comet assay was performed using a modified version of the neutral comet protocol as previously described.Citation22 Briefly, gels were lysed for 4 h at 43°C (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris, 1% N-Lauroylsarcosine, pH 9.5 with 0.5% Triton X-100 and 10% DMSO added 20 min before use). After lysis the slides were washed three times for 30 min with the electrophoresis buffer (90 mM Tris, 90 mM Boric Acid, 2 mM Na2EDTA, pH 8.5). Electrophoresis was conducted at 4°C for 1 h at 0.6 V/cm and 6 mA.
Fluorescence imaging and comet analysis
After electrophoresis, gels were neutralized in 0.4 M Tris, pH 7.5 (3 × 5 min). Slides were then stained with SYBR Gold (Invitrogen). Images were captured automatically using an epifluorescent microscope and analyzed automatically using custom software written in MATLAB (The Mathworks). Traditional comet slides were scored manually using Komet 5.5 (Andor Technology). ANOVA analysis was conducted to determine statistical significance between irradiation doses () and a Student’s t-test was used to evaluate relative repair differences between cells (–).
γ-H2AX Assay
Wild type (CHO-K1) and DNA-PKcs−/− (irs-20) CHO cells were plated in 6-well plates and allowed to grow to confluency over night. Media containing 50 μM NU7441 inhibitor was added to wild type cells for 1 h at 37°C, 5% CO2. All plates except the untreated control were exposed to 100 Gy in the Cobalt-60 irradiator (Gammacell 220 Excel, MDS Nordion). Cells were allowed to repair for (0, 1, 2, 4, 8 h) in either fresh medium or medium supplemented with NU7441. Cell samples were lysed at each time point [0.12 M TRIS-HCl pH 6.8 with Photostop tab (Roche Cat. #04906837001) and complete Mini (Roche Cat. #05056489001)], scraped from the dish and stored in eppendorf tubes for γ-H2AX analysis by western blot. Briefly, samples were sonicated and run in the high-throughput E-Page gel system (Invitrogen) and transferred using the iBlot Blotting System (Invitrogen). γ-H2AX was visualized using anti-γ-H2AX rabbit monoclonal antibody (Cat. #9718, Cell Signaling Technology) and normalized to actin levels using anti-actin mouse monoclonal antibody (Cat. #A5441, Cell Signaling Technology).
| Abbreviations: | ||
| DSB | = | double-strand break |
| NHEJ | = | non-homologous endjoining |
| IR | = | ionizing radiation |
| HR | = | homologous recombination |
| DNA-PK | = | DNA protein kinase |
| PI3K | = | phosphatidylinositol 3-kinase |
| PI3KKs | = | phosphatidylinositol 3-kinase related kinases |
Additional material
Download Zip (85 KB)Acknowledgment
This work was supported primarily by U01-ES016045 with partial support from R21-ES019498, ES015339 and CA112967. Equipment for data analysis was provided by the Center for Environmental Health Sciences P30-ES002109. D.M.W. was supported by the NIEHS Training Grant in Environmental Toxicology T32-ES007020. We thank Dwight Chambers for his valuable contributions. We also thank Joel Bedford and Hatsumi Nagasawa for the irs-20 cell line and Larry Thompson for CHO cell lines.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/23880
References
- Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer 2002; 2:188 - 200; http://dx.doi.org/10.1038/nrc749; PMID: 11990855
- Bernier J, Hall EJ, Giaccia A. Radiation oncology: a century of achievements. Nat Rev Cancer 2004; 4:737 - 47; http://dx.doi.org/10.1038/nrc1451; PMID: 15343280
- Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, et al. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet Genome Res 2004; 104:14 - 20; http://dx.doi.org/10.1159/000077461; PMID: 15162010
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007; 6:923 - 35; http://dx.doi.org/10.1016/j.dnarep.2007.02.006; PMID: 17363343
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res 2008; 18:134 - 47; http://dx.doi.org/10.1038/cr.2007.111; PMID: 18157161
- Dikomey E, Dahm-Daphi J, Brammer I, Martensen R, Kaina B. Correlation between cellular radiosensitivity and non-repaired double-strand breaks studied in nine mammalian cell lines. Int J Radiat Biol 1998; 73:269 - 78; http://dx.doi.org/10.1080/095530098142365; PMID: 9525255
- Tan DS-W, Gerlinger M, Teh B-T, Swanton C. Anti-cancer drug resistance: understanding the mechanisms through the use of integrative genomics and functional RNA interference. Eur J Cancer 2010; 46:2166 - 77; http://dx.doi.org/10.1016/j.ejca.2010.03.019; PMID: 20413300
- Madhusudan S, Hickson ID. DNA repair inhibition: a selective tumour targeting strategy. Trends Mol Med 2005; 11:503 - 11; http://dx.doi.org/10.1016/j.molmed.2005.09.004; PMID: 16214418
- Ding J, Miao ZH, Meng LH, Geng MY. Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol Sci 2006; 27:338 - 44; http://dx.doi.org/10.1016/j.tips.2006.04.007; PMID: 16697054
- Curtin N. Therapeutic potential of drugs to modulate DNA repair in cancer. Expert Opin Ther Targets 2007; 11:783 - 99; http://dx.doi.org/10.1517/14728222.11.6.783; PMID: 17504016
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 2008; 8:193 - 204; http://dx.doi.org/10.1038/nrc2342; PMID: 18256616
- Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008; 36:5678 - 94; http://dx.doi.org/10.1093/nar/gkn550; PMID: 18772227
- An J, Huang Y-C, Xu Q-Z, Zhou L-J, Shang Z-F, Huang B, et al. DNA-PKcs plays a dominant role in the regulation of H2AX phosphorylation in response to DNA damage and cell cycle progression. BMC Mol Biol 2010; 11:18; http://dx.doi.org/10.1186/1471-2199-11-18; PMID: 20205745
- Kohn KW, Grimek-Ewig RA. Alkaline elution analysis, a new approach to the study of DNA single-strand interruptions in cells. Cancer Res 1973; 33:1849 - 53; PMID: 4737199
- Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006; 1:23 - 9; http://dx.doi.org/10.1038/nprot.2006.5; PMID: 17406208
- Van Kooij RJ, de Boer P, De Vreeden-Elbertse JM, Ganga NA, Singh N, Te Velde ER. The neutral comet assay detects double strand DNA damage in selected and unselected human spermatozoa of normospermic donors. Int J Androl 2004; 27:140 - 6; http://dx.doi.org/10.1111/j.1365-2605.2004.00463.x; PMID: 15139968
- Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun 1984; 123:291 - 8; http://dx.doi.org/10.1016/0006-291X(84)90411-X; PMID: 6477583
- Fairbairn DW, Olive PL, O’Neill KL. The comet assay: a comprehensive review. Mutat Res 1995; 339:37 - 59; http://dx.doi.org/10.1016/0165-1110(94)00013-3; PMID: 7877644
- Collins AR, Oscoz AA, Brunborg G, Gaivão I, Giovannelli L, Kruszewski M, et al. The comet assay: topical issues. Mutagenesis 2008; 23:143 - 51; http://dx.doi.org/10.1093/mutage/gem051; PMID: 18283046
- Belyaev IY, Eriksson S, Nygren J, Torudd J, Harms-Ringdahl M. Effects of ethidium bromide on DNA loop organisation in human lymphocytes measured by anomalous viscosity time dependence and single cell gel electrophoresis. Biochim Biophys Acta 1999; 1428:348 - 56; http://dx.doi.org/10.1016/S0304-4165(99)00076-8; PMID: 10434054
- García O, Mandina T, Lamadrid AI, Diaz A, Remigio A, Gonzalez Y, et al. Sensitivity and variability of visual scoring in the comet assay. Results of an inter-laboratory scoring exercise with the use of silver staining. Mutat Res 2004; 556:25 - 34; PMID: 15491629
- Forchhammer L, Johansson C, Loft S, Möller L, Godschalk RWL, Langie SAS, et al. Variation in the measurement of DNA damage by comet assay measured by the ECVAG inter-laboratory validation trial. Mutagenesis 2010; 25:113 - 23; http://dx.doi.org/10.1093/mutage/gep048; PMID: 19910383
- Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci USA 2010; 107:10008 - 13; http://dx.doi.org/10.1073/pnas.1004056107; PMID: 20534572
- Wojewódzka M, Buraczewska I, Kruszewski M. A modified neutral comet assay: elimination of lysis at high temperature and validation of the assay with anti-single-stranded DNA antibody. Mutat Res 2002; 518:9 - 20; http://dx.doi.org/10.1016/S1383-5718(02)00070-0; PMID: 12063063
- Nichols C, Kollmannsberger C. First-line chemotherapy of disseminated germ cell tumors. Hematol Oncol Clin North Am 2011; 25:543 - 56, viii; http://dx.doi.org/10.1016/j.hoc.2011.03.011; PMID: 21570608
- Richardson SE, McNamara C. The Management of Classical Hodgkin’s Lymphoma: Past, Present, and Future. Adv Hematol 2011; 2011:865870; http://dx.doi.org/10.1155/2011/865870; PMID: 21687653
- Linnert M, Gehl J. Bleomycin treatment of brain tumors: an evaluation. Anticancer Drugs 2009; 20:157 - 64; http://dx.doi.org/10.1097/CAD.0b013e328325465e; PMID: 19396014
- Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res 2000; 28:2585 - 96; http://dx.doi.org/10.1093/nar/28.13.2585; PMID: 10871410
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem 2010; 285:1414 - 23; http://dx.doi.org/10.1074/jbc.M109.065615; PMID: 19893054
- Ismail IH, Mårtensson S, Moshinsky D, Rice A, Tang C, Howlett A, et al. SU11752 inhibits the DNA-dependent protein kinase and DNA double-strand break repair resulting in ionizing radiation sensitization. Oncogene 2004; 23:873 - 82; http://dx.doi.org/10.1038/sj.onc.1207303; PMID: 14661061
- Losada R, Rivero MT, Slijepcevic P, Goyanes V, Fernández JL. Effect of Wortmannin on the repair profiles of DNA double-strand breaks in the whole genome and in interstitial telomeric sequences of Chinese hamster cells. Mutat Res 2005; 570:119 - 28; http://dx.doi.org/10.1016/j.mrfmmm.2004.10.009; PMID: 15680409
- Perrault R, Wang H, Wang M, Rosidi B, Iliakis G. Backup pathways of NHEJ are suppressed by DNA-PK. J Cell Biochem 2004; 92:781 - 94; http://dx.doi.org/10.1002/jcb.20104; PMID: 15211575
- Takahashi A, Ohnishi T. Does gammaH2AX foci formation depend on the presence of DNA double strand breaks?. Cancer Lett 2005; 229:171 - 9; http://dx.doi.org/10.1016/j.canlet.2005.07.016; PMID: 16129552
- Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene 2005; 24:949 - 61; http://dx.doi.org/10.1038/sj.onc.1208332; PMID: 15592499
- Hosoi Y, Miyachi H, Matsumoto Y, Ikehata H, Komura J, Ishii K, et al. A phosphatidylinositol 3-kinase inhibitor wortmannin induces radioresistant DNA synthesis and sensitizes cells to bleomycin and ionizing radiation. Int J Cancer 1998; 78:642 - 7; http://dx.doi.org/10.1002/(SICI)1097-0215(19981123)78:5<642::AID-IJC19>3.0.CO;2-3; PMID: 9808536
- Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 1994; 269:5241 - 8; PMID: 8106507
- Hollick JJ, Golding BT, Hardcastle IR, Martin N, Richardson C, Rigoreau LJM, et al. 2,6-disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-Dependent protein kinase (DNA-PK). Bioorg Med Chem Lett 2003; 13:3083 - 6; http://dx.doi.org/10.1016/S0960-894X(03)00652-8; PMID: 12941339
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 2004; 14:6083 - 7; http://dx.doi.org/10.1016/j.bmcl.2004.09.060; PMID: 15546735
- Griffin RJ, Fontana G, Golding BT, Guiard S, Hardcastle IR, Leahy JJ, et al. Selective benzopyranone and pyrimido[2,1-a]isoquinolin-4-one inhibitors of DNA-dependent protein kinase: synthesis, structure-activity studies, and radiosensitization of a human tumor cell line in vitro. J Med Chem 2005; 48:569 - 85; http://dx.doi.org/10.1021/jm049526a; PMID: 15658870
- Kashishian A, Douangpanya H, Clark D, Schlachter ST, Eary CT, Schiro JG, et al. DNA-dependent protein kinase inhibitors as drug candidates for the treatment of cancer. Mol Cancer Ther 2003; 2:1257 - 64; PMID: 14707266
- Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res 2006; 66:5354 - 62; http://dx.doi.org/10.1158/0008-5472.CAN-05-4275; PMID: 16707462
- Chen H, Ma Z, Vanderwaal RP, Feng Z, Gonzalez-Suarez I, Wang S, et al. The mTOR inhibitor rapamycin suppresses DNA double-strand break repair. Radiat Res 2011; 175:214 - 24; http://dx.doi.org/10.1667/RR2323.1; PMID: 21268715
- Kiskinis E, Suter W, Hartmann A. High throughput Comet assay using 96-well plates. Mutagenesis 2002; 17:37 - 43; http://dx.doi.org/10.1093/mutage/17.1.37; PMID: 11752232
- Priestley A, Beamish HJ, Gell D, Amatucci AG, Muhlmann-Diaz MC, Singleton BK, et al. Molecular and biochemical characterisation of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res 1998; 26:1965 - 73; http://dx.doi.org/10.1093/nar/26.8.1965; PMID: 9518490
- Izzard RA, Jackson SP, Smith GC. Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res 1999; 59:2581 - 6; PMID: 10363977
- Kumar A, Fernandez-Capetillo O, Carrera AC, Carrera AC. Nuclear phosphoinositide 3-kinase beta controls double-strand break DNA repair. Proc Natl Acad Sci USA 2010; 107:7491 - 6; http://dx.doi.org/10.1073/pnas.0914242107; PMID: 20368419
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene 2008; 27:5497 - 510; http://dx.doi.org/10.1038/onc.2008.245; PMID: 18794884
- Mazzoletti M, Bortolin F, Brunelli L, Pastorelli R, Di Giandomenico S, Erba E, et al. Combination of PI3K/mTOR inhibitors: antitumor activity and molecular correlates. Cancer Res 2011; 71:4573 - 84; http://dx.doi.org/10.1158/0008-5472.CAN-10-4322; PMID: 21602434
- Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 2007; 5:981 - 9; http://dx.doi.org/10.1158/1541-7786.MCR-07-0324; PMID: 17951399