Abstract
Chk1 is implicated in several checkpoints of the cell cycle acting as a key player in the signal transduction pathway activated in response to DNA damage and crucial for the maintenance of genomic stability. Chk1 also plays a role in the mitotic spindle checkpoint, which ensures the fidelity of mitotic segregation during mitosis, preventing chromosomal instability and aneuploidy. Mad2 is one of the main mitotic checkpoint components and also exerts a role in the cellular response to DNA damage. To investigate a possible crosslink existing between Chk1 and Mad2, we studied Mad2 protein levels after Chk1 inhibition either by specific siRNAs or by a specific and selective Chk1 inhibitor (PF-00477736), and we found that after Chk1 inhibition, Mad2 protein levels decrease only in tumor cells sensitive to Chk1 depletion. We then mapped six Chk1’s phosphorylatable sites on Mad2 protein, and found that Chk1 is able to phosphorylate Mad2 in vitro on more than one site, while it is incapable of phoshorylating the Mad2 form mutated on all six phosphorylatable sites. Moreover our studies demonstrate that Chk1 co-localizes and physically associates with Mad2 in cells both under unstressed conditions and after DNA damage, thus providing new and interesting evidence on Chk1 and Mad2 crosstalk in the DNA damage checkpoint and in the mitotic spindle checkpoint.
Introduction
A series of highly regulated and coordinated processes ensures that chromosomes are reproduced and equally distributed to daughter cells in each cell division cycle. The mitotic spindle checkpoint guarantees the fidelity of mitotic segregation during mitosis, preventing chromosomal instability and aneuploidy, events that contribute to malignant transformation.Citation1-Citation3 This checkpoint is active during each cell cycle, until all the chromosomes are well aligned on the metaphase plate, and the kinetochores are attached to mitotic spindle. During activation of the mitotic spindle checkpoint, its main components (Mad2, BubR1, Mad1) form an inhibitory ternary complex with an E3 ligase, the anaphase-promoting complex (APC) and its substrate-specific activator, Cdc20.Citation4,Citation5 After the bipolar attachments of all the chromosomes to the mitotic spindle at metaphase, the multi-protein mitotic checkpoint complex is extinguished, leading to activation of Cdc20, which, in turn, influences a series of molecular events ending with chromosomes separation.Citation6,Citation7
Chk1 is a conserved protein kinase, originally identified in fission yeast, required to delay entry of cells with damaged or unreplicated DNA into mitosis.Citation8,Citation9 The requirement of Chk1 for both S and G2/M checkpoints has been clarified in the last few years, and its role in sustaining activation of the spindle checkpoint whenever cells with damaged DNA enter mitosis has been described in fission and budding yeast.Citation10,Citation11 Chk1-deficient human cells fail to sustain mitotic arrest after treatment with spindle poisons. Chk1-dependent BubR1 phosphorylation through Aurora B was found after taxol treatment in lymphoma DT40 cells and after Nocodazole (Noc) in U2OS cells.Citation12,Citation13 Haploid loss of Chk1 impaired the spindle checkpoint activated by Noc in mouse mammary epithelial cells and in mammary cancer cells, providing further evidence of its role in this checkpoint.Citation14 Chk1 downregulation in U2OS cells also led to lower protein levels of Mad2.Citation13 The lack of Mad2 in human cells leads to mitotic checkpoint inactivation and chromosomal instability.Citation15 Mad2, like Chk1, is essential for mouse embryogenesis, as Mad2−/− KO mice die at E6.5 as a consequence of chromosome missegregation and apoptosis.Citation16 These data suggest that Chk1 and Mad2 may be involved in the same pathway regulating the accurate progression of mitosis, and Mad2 may be important for Chk1 function in the spindle checkpoint. Experimental evidence supports this relationship in fission yeast, where the Chk1-dependent delay of metaphase-anaphase transition in mitotic cells with damaged DNA was Mad2-dependent.Citation10 A link has been suggested between mitotic checkpoint and DNA damage response in yeast and mammalsCitation17,Citation18 and Mad2 is a key factor not only in maintaining genomic stability, but also in regulating cellular sensitivity to certain anticancer drugs (such as cis-platinum).Citation19,Citation20 It is still not clear how Chk1 and Mad2 are functionally connected. We herein report new evidence that corroborates a relationship between Chk1 and Mad2, providing an additional crosslink between the mitotic spindle and DNA damage checkpoints.
Results and Discussion
Chk1 inhibition lowers protein levels of Mad2 in cells sensitive to Chk1 depletion
Our previous data demonstrated that Chk1 downregulation by siRNA in U2OS cells induced cell death as a consequence of aberrant mitosis and defects in the activation of the mitotic spindle checkpoint. In these conditions, decreased levels of the mitotic checkpoint protein Mad2 were observed.Citation13 To further investigate the Chk1-dependent regulation of Mad2 protein, we investigated a wider panel of cancer cell lines. The ovarian cancer cell lines OVCAR-8, OVCA432, A2780 and OVCAR-5 were transiently transfected with either scramble or Chk1 siRNAs. The U2OS osteosarcoma cell line was included as positive control. Chk1 and Mad2 protein levels () and cell survival () were evaluated 72 h after transfection. The decrease in Mad2 protein levels after Chk1 depletion correlates with the extent of the effect of Chk1 downregulation on cell survival. In fact, in the cell lines with approximately 60% compromised survival after Chk1 depletion (U2OS, OVCAR-8 and OVCA-432), the decrease in Mad2 protein levels ranged from 40 to 60%, while in cell lines whose survival was not affected (OVCAR-5 and A2780) Mad2 protein levels were unchanged. To see whether in U2OS, OVCAR-8 and OVCA-432, the drop in Mad2 protein was related to a decrease in mRNA, Mad2 mRNA levels were measured 72 h after siRNAs transfection. Mad2 mRNA levels did not fall (). These data fit well with our previous observation in U2OS cells, and partially with recent data, that the haploid loss of Chk1 in tumor lines derived from mice mammary glands and susceptible to Chk1 loss correlated not only with low levels of protein, but also with low mRNA levels of the mitotic checkpoint component Mad2.Citation14 This difference may be due to the different experimental systems used (Chk1+/− cell lines) and possibly to the extent of Chk1 downregulation and/or inhibition. Studies on Mad2 protein stability in U2OS cells transfected with Chk1 siRNA after treatment with cycloheximide (CHX) further suggested that Mad2 downregulation is occurring at protein level in this experimental condition (). In fact, after treatment with CHX, Mad2 protein was more unstable in Chk1 siRNA-transfected sample starting from 3 h after treatment with CHX as compared with scramble siRNA-transfected cells. In addition, the effect of Mad2 depletion on cell survival was studied in U2OS cells, and, as shown in Figure S1A, Mad2 protein levels were downregulated at 72 and 144 h after transfection with a specific siRNA against Mad2, and, interestingly, at a 144 h time point a concurrent decrease in Chk1 protein levels was observed. Mad2 downregulation caused an inhibition of cell growth evident at 144 h after transfection (Fig. S1B), a time point at which Chk1 protein was also found at very low levels, rendering difficult to discriminate the effect of each protein. However, these data strongly suggest that the two proteins may be physically interacting in this experimental system.
Figure 1. (A) Western blot analysis of Chk1, Mad2 and Actin in U2OS, OVCAR8, OVCA432, A2780 and OVCAR-5 cells 72 h after either scramble or Chk1 siRNA transfection. (B) Densitometric analysis of Mad2 protein levels in the cell lines 72 h after siRNA Chk1 transfection, compared with the siRNA scramble transfected cells. Data are percentages of siRNA scramble controls, representing the ratio of Mad2 to actin. (C) Schematic representation of the percentage of cells surviving at 72 h after transfection with siRNA Chk1. Data are expressed as the percentages of scramble transfected cells and are the mean ± SD of two independent experiments.
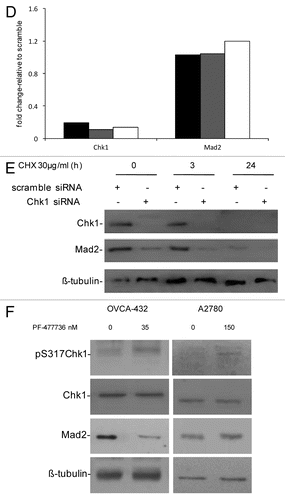
Figure 1D–F. (D) Chk1 and Mad2 mRNA expression in U2OS (black bars), OVCAR-8 (gray bars) and OVCA-432 (white bars) by real-time PCR 72 h after transfection with siRNA Chk1. Chk1 and Mad2 expression levels are normalized to the internal mRNA levels of actin and are represented as the fold change from scramble transfected samples. (E) Western blot analysis of Chk1, Mad2 and β-tubulin in U2OS cells transfected with either scramble or Chk1 siRNA and 72 h post-transfection either untreated or treated with CHX 30 µg/ml for 3 and 24 h. (F) Western blot analysis of Mad2, pS317-Chk1, Chk1 and β-tubulin in OVCA-432 and A2780 cells 72 h after treatment with 35 or 150 nM of the Chk1 inhibitor PF-00477736 compared with untreated cells.
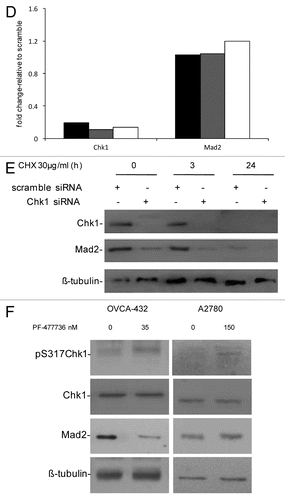
The OVCA-432 cell line is more sensitive to a selective and specific Chk1 inhibitor, PF-00477736, than other cancer cell lines (e.g., A2780).Citation21 shows that Mad2 protein levels decrease in OVCA-432 cells treated with 35 nM PF-00477736 but did not change in A2780 cells, which are more resistant to Chk1 inhibition, after 150 nM PF-00477736. In both cell lines, Chk1 kinase activity was inhibited, as shown by increased phosphorylation in S317 of Chk1 after treatment with PF-00477736, bona fide for Chk1 inhibition.Citation22 Recent data suggest that Chk1 inhibitors could also be used as single agents in a subset of cancers, so the decrease in Mad2 protein levels only in the presence of a cytotoxic effect could be considered a pharmacodynamic read-out of the susceptibility of tumor cells to Chk1 inhibitors. These data are in agreement with those obtained with siRNA Chk1 and further indicate that the Chk1 kinase activity per se is sufficient to cause this effect; therefore, Chk1 kinase might induce post-translational modifications of Mad2 important for its stability. A previous study found that Mad2 is subjected to post-translational modification by phosphorylation, though little is known about its biological significance. Mad2 phosphorylation mostly occurs in the C-terminal region of the protein in mitotic cells and regulates its checkpoint activity by modulating its association with Mad1 and the APC.Citation23
Chk1 protein kinase phosphorylates Mad2 in vitro
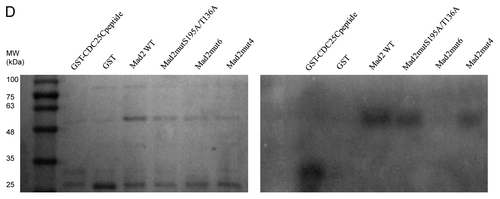
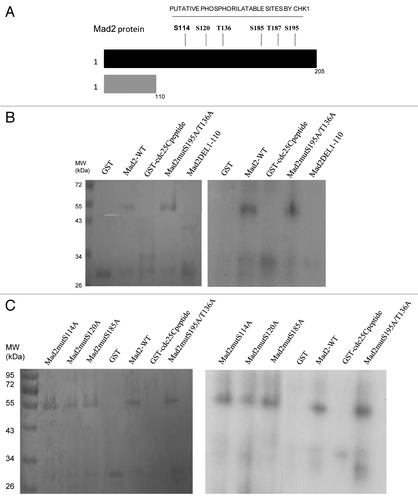
To further investigate a possible relationship between Chk1 and Mad2, the Mad2 protein sequence was investigated to check for putative Chk1 phosphorylatable sites. The minimal Chk1 phosphorylatable consensus site consists of the following motif: β-X-X-(S/T), where X can be any aminoacid residue, and β should be a basic aminoacid, especially either R or K.Citation24,Citation25 The full consensus site consists of the extended motif Φ-X-β-X-X-(S/T)-Φ, where additionally Φ in position −5 and +1 has to be a hydrophobic residue [i.e., MFLVI(A)], which has an important anchoring function to the kinase catalytic pocket according to the crystal Chk1 structure.Citation24,Citation26 Analyzing the Mad2 protein sequence, we detected two sites, T136 and S195, contained in a full consensus site and four other sites included in the minimal Chk1 phosphorylatable consensus site: S114, S120, S185 and T187 (). An in vitro kinase assay with purified recombinant Chk1 and Mad2 human proteins showed that Chk1 phosphorylates Mad2 in vitro (). To better define the regions and sites of Mad2 phosphorylatable by Chk1, Mad2-site specific point mutations and deleted regions were produced as recombinant proteins (). The C-terminally truncated Mad2 protein 1–110 lacking all the Chk1 phosphorylatable sites was not phosphorylated in vitro by Chk1 (), while the Mad2-GST recombinant protein double-point-mutated in S195 and T136 sites was phosphorylated. Similarly, the single-point-mutated Mad2 proteins, S114A, S120A, S185A and T187A, were phosphorylated in vitro by Chk1 ( and data not shown). Finally the Mad2-GST recombinant protein mutated in all the six phosphorylatable sites could not be phosphorylated in vitro by Chk1, while the mutated form in four phosphorylatable sites (S114A, S120A, T136 and S195) is still phosphorylated (). The discovery of the kinases involved in phosphorylation of Mad2 may be important for a better understanding of the mitotic checkpoint pathway. Mad2 can be phosphorylated in vitro by Nek2, and this phosphorylation is important for its dimeric conformation. The Mad2 N-terminal region seems to be involved in this translational modification (presumably T136), while the C-terminal region (which is associated with other checkpoint proteins such as Mad1 and Cdc20) is not required. The authors showed that Nek2 cooperates with Mad2 in negatively regulating the APC.Citation27 Our data indicate that Chk1 can phosphorylate Mad2 in vitro in more than one site, as it could still phosphorylate all the specific single-point mutations and the double mutant S195A and T136A. The fact that the mutant form in the four phosphorylatable sites (S114A, S120A, T136 and S195) is still phosphorylated, while Mad2 mutated form in all the phosphorylatable sites could not, further suggests that S185 and/or T187 are the main candidates for the phosphorylation by Chk1. The functional significance of the phosphorylated-Chk1-induced form of Mad2 in vivo is far to be elucidated, and the lack of commercially available specific phospho Mad2 antibodies, at least at present, is a limiting factor.
Figure 2. (A) Schematic representation of the Chk1 phosphorylatable sites on the Mad2 protein sequence. Specific S/T sites are evidenced in bold type. The boxes illustrate the two complete Chk1 phosphorylatable sites of Mad2. (B) In vitro kinase assay using wt recombinant GST-Chk1 in the presence of 32γ ATP and the positive recombinant substrate GST-Cdc25C, the putative recombinant substrate GST-Mad2 and the negative control GST. Left: SDS-PAGE stained with comassie blue. Right: autoradiography.
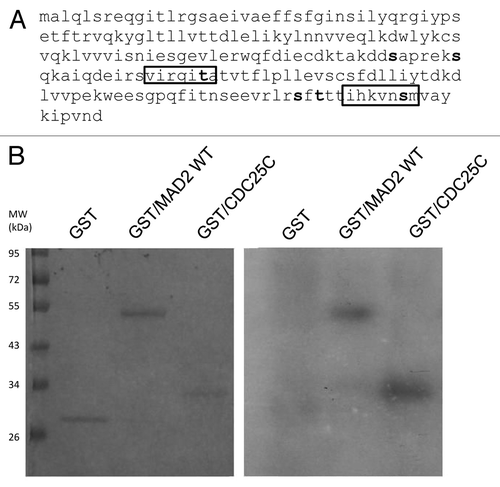
Figure 3. A Schematic representation of the Mad2 protein region with Chk1 phosphorylatable sites and the C-terminally truncated part with no phosphorylatable sites (1–110). (B–D) In vitro kinase assay using wt recombinant GST-Chk1 in the presence of 32γ ATP and the positive recombinant substrate GST-Cdc25C, the putative recombinant substrate GST-Mad2 wt, the mutated proteins S195A/T136A and the DEL1–110 (B), the mutated proteins S114A, S120A and S185A (C) and the mutated form in 4 and 6 phosphorylatable sites (D). GST, negative control. Left: SDS-PAGE stained with comassie blue. Right: autoradiography.
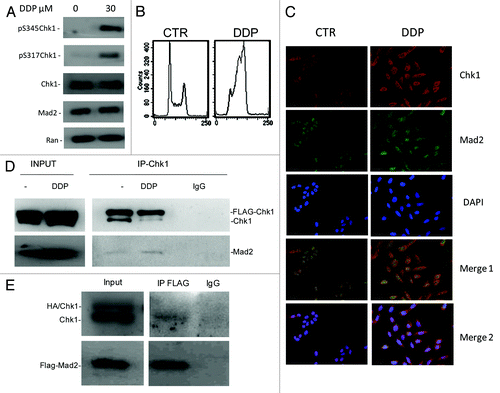
Mad2 co-localization and association with Chk1 increase after DDP treatment
To further investigate the Chk1/Mad2 interaction in cells, we co-stained Chk1 and Mad2 in U2OS cells under unstressed conditions and 24 h after treatment with DDP 30 µM (a concentration close to the IC50 for U2OS cells and able to induce Chk1 activation). Mad2 and Chk1 have both been recently described as involved in the DNA damage response besides their role in the mitotic spindle checkpoint.Citation20 U2OS treatment with DDP induced Chk1 activation (increased phosphorylation at sites S345 and S317) with no detectable changes in Mad2 levels () and a clear G2 block of the cell cycle (). Mad2 and Chk1 co-localized mostly in the nucleus in interphase cells under unstressed conditions, as already described ().Citation28,Citation29 After DDP treatment, the co-localization of the two proteins slightly increased, and there was clear redistribution in the perinuclear region (; and white arrows in merge blow ups in Fig. S2). With DNA damage, the phosphorylated and activated form of Chk1 is released from chromatin into the soluble nucleus and later to the cytoplasm.Citation28 The scanty data on Mad2 localization after DNA damage suggests that it localizes in the nucleopore complex and in the cytoplasm, with a role in the intracellular redistribution of repair proteins in response to DNA damage, as for NER proteins.Citation20 To determine the potential physical association between Mad2 and Chk1, endogenous and exogenous Chk1 were immunoprecipitated from cellular extracts of U2OS cells overexpressing both Flag-Chk1 and Mad2, either untreated or treated with DDP and then immunoblotted for Mad2. Mad2 co-immunoprecipitated with Chk1 from cellular extracts of untreated cells, and this association slightly increased after treatment with DDP (). Chk1 also co-immunoprecipitated with anti-Flag in U2OS cells overexpressing Flag-Mad2 (). We have herein showed that Chk1 and Mad2 not only co-localize, but also physically associate and the co-localization and interaction slightly increase after DNA damage. The co-localization, mostly occurring in the perinuclear area in the cytoplasm, suggests that the two proteins may cooperate in regulating the DNA damage response. However, the re-distribution of the two proteins in the perinuclear area may also be a consequence of the accumulation of cells in G2 phase (). In U2OS cells enriched in G2 phase after treatment with aphidicolin, there was a clear co-localization of Chk1 and Mad2 in the perinuclear area (Fig. S3). Before cells enter mitosis at the G2-M transition, the increased activity of CDK1/CyclinB leads to Chk1 phosphorylation and subsequent accumulation in the cytoplasm.Citation30 The co-localization in the cytoplasm of Chk1 and Mad2 may be essential for Mad2 to exert its checkpoint function during the metaphase-anaphase transition, but further studies are needed to elucidate the significance of Chk1-Mad2 association and co-localization after DDP treatment. In conclusion, these findings point to the existence of cross-talk between Chk1 and Mad2. In addition, a decrease in Mad2 protein levels was found to be a possible biomarker of cellular sensitivity to Chk1 inhibition/depletion. We show for the first time that Chk1 can phosphorylate Mad2 in vitro and can associate and co-localize with Mad2 in cells in unstressed conditions and after DNA damage. The meaning of this interaction remains to be defined. Chk1/Mad2 cross-talk might have a key role in both the DNA damage and the mitotic spindle checkpoints.
Figure 4. (A) Western blot analysis of pS345-Chk1, pS317-Chk1, Chk1, Mad2 and Ran in U2OS cells 24 h after treatment with 30 µM of DDP and in untreated cells. (B) Analysis of DNA content by FACS 24 h after the end of treatment with 30 µM of DDP and in untreated samples. (C) Chk1, Mad2 and DAPI staining in U2OS cells. Merge 1: Mad2 and Chk1 overlay. Merge 2: Chk1, Mad2 and DAPI overlay. (D) Exogenous (Flag-Chk1) and endogenous Chk1 immunoprecipitation from cell extracts of U2OS overexpressing Flag-Chk1 and Mad2, either untreated or 24 h after treatment with 30 µM DDP using a rabbit polyclonal Chk1 antibody. Immunoprecipitation with Rabbit IgG: negative control of the experiment. Immunoprecipitates probed by western blot analysis for Chk1 and Mad2 protein levels. Inputs: total cell extracts, either untreated or treated with DDP (the experiment showed is representative of two independent experiments). (E) Exogenous Flag-Mad2 immunoprecipitation from cell extracts of U2OS overexpressing Flag-Mad2 and HA-Chk1, using a mouse monoclonal Flag antibody. Immunoprecipitation with Mouse IgG: negative control of the experiment. Immunoprecipitates probed by western blot analysis for Chk1 and Mad2 protein levels. Inputs: total cell extracts.
Materials and Methods
Cell lines, transfections and drugs
Information on cell lines, transfection procedures and drugs are available in the Supplemental Materials packet.
Western blotting analysis
Proteins were extracted and visualized using standard techniques, and as already described.Citation13 Primary anti Chk1 (G4), Mad2 (FL-205), β-tubulin (H-235), actin (C-11) were purchased from Santa Cruz Biotechnology. The mouse monoclonal anti Ran (clone 20) was from BD Transduction Laboratories. Primary anti pS317- and pS345-Chk1 were purchased from Cell Signaling Technology. Densitometric Analysis was done with ImageJ.
RNA isolation, cDNA preparation and real-time PCR
Total RNA was purified using the SV-Total RNA isolation system (Promega). One microgram of total RNA was reverse-transcribed to cDNA in 20 µL of reaction mix with an Archive Kit (Applied Biosystems) using random primers. Real-time PCR was run to quantify Chk1, Mad2 and actin. Primers were synthesized by Sigma. Real-time PCR was done using the 7900HT Sequence Detection System (Applied Biosystems).
Purification of GST-fusion proteins and protein kinase assay
Chk1 kinase activity was determined after isolation of recombinant Chk1 fused to GST from bacteria. Substrates were obtained as recombinant protein from bacteria as well. Details are described in Supplemental Materials.
Direct-site specific mutagenesis and deleted forms of Mad2
To obtain Mad2 proteins with point mutations in the phosphorylatable sites, S114, S120, S185, S195 and T136 were substituted with alanine (non-polar aminoacid) sequentially. The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used, following the manufacturer’s instructions. To obtain the deleted forms of Mad2, ad hoc PCR programs were used to amplify specific Mad2 cDNA regions and molecular cloning permitted the construction of pGEX-3X/GST-constructs with the deleted forms of the protein of interest.
Immunofluorescence
Specifications of Immunofluorescence staining are provided in Supplemental Materials.
Immunoprecipitation
The experiment to assess the co-immunoprecipitation of Mad2 with Chk1 in U2OS cells is described in Supplemental Materials.
FACS analysis
To detect DNA by FACS, cells were fixed 24 h after the end of treatment with DDP or 16 h after the release from aphidicolin; they were washed twice in ice-cold PBS, fixed in ice-cold 70% ethanol, washed in PBS, resuspended in 2 mL of a solution containing 25 μg/mL of propidium iodide in PBS and 25 μL of RNase 1 mg/mL in water, and stained overnight at 4°C in the dark. Cell cycle analysis was done on at least 10,000 cells for each sample using the FACS Calibur (Becton Dickinson).
Additional material
Download Zip (600.6 KB)Acknowledgments
The generous contribution of the Italian Association for Cancer Research (AIRC), Milan, Italy, is gratefully acknowledged. We thank J.D. Baggott that kindly edited the paper. Thanks to Alessandra Basana, an undergraduate student that helped with interest and passion while she was learning how to work in a laboratory. A special thanks to Faina Vikhanskaya, who has always pushed me up in performing one more additional recombinant protein extraction and kinase assay after every bad result. She will always be a great example of scientist to me.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/24090
References
- Suijkerbuijk SJ, Kops GJ. Preventing aneuploidy: the contribution of mitotic checkpoint proteins. Biochim Biophys Acta 2008; 1786:24 - 31; PMID: 18472014
- Draviam VM, Xie S, Sorger PK. Chromosome segregation and genomic stability. Curr Opin Genet Dev 2004; 14:120 - 5; http://dx.doi.org/10.1016/j.gde.2004.02.007; PMID: 15196457
- Laiho M, Latonen L. Cell cycle control, DNA damage checkpoints and cancer. Ann Med 2003; 35:391 - 7; http://dx.doi.org/10.1080/07853890310014605; PMID: 14572162
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev 1998; 12:1871 - 83; http://dx.doi.org/10.1101/gad.12.12.1871; PMID: 9637688
- Wassmann K, Benezra R. Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc Natl Acad Sci USA 1998; 95:11193 - 8; http://dx.doi.org/10.1073/pnas.95.19.11193; PMID: 9736712
- Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell 2002; 9:931 - 43; http://dx.doi.org/10.1016/S1097-2765(02)00540-3; PMID: 12049731
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell 2007; 27:3 - 16; http://dx.doi.org/10.1016/j.molcel.2007.06.009; PMID: 17612486
- Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 1993; 363:368 - 71; http://dx.doi.org/10.1038/363368a0; PMID: 8497322
- Zachos G, Rainey MD, Gillespie DA. Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol Cell Biol 2005; 25:563 - 74; http://dx.doi.org/10.1128/MCB.25.2.563-574.2005; PMID: 15632059
- Collura A, Blaisonneau J, Baldacci G, Francesconi S. The fission yeast Crb2/Chk1 pathway coordinates the DNA damage and spindle checkpoint in response to replication stress induced by topoisomerase I inhibitor. Mol Cell Biol 2005; 25:7889 - 99; http://dx.doi.org/10.1128/MCB.25.17.7889-7899.2005; PMID: 16107732
- Searle JS, Schollaert KL, Wilkins BJ, Sanchez Y. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat Cell Biol 2004; 6:138 - 45; http://dx.doi.org/10.1038/ncb1092; PMID: 14743219
- Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, et al. Chk1 is required for spindle checkpoint function. Dev Cell 2007; 12:247 - 60; http://dx.doi.org/10.1016/j.devcel.2007.01.003; PMID: 17276342
- Carrassa L, Sanchez Y, Erba E, Damia G. U2OS cells lacking Chk1 undergo aberrant mitosis and fail to activate the spindle checkpoint. J Cell Mol Med 2009; 13:8A 1565 - 76; PMID: 19778378
- Fishler T, Li YY, Wang RH, Kim HS, Sengupta K, Vassilopoulos A, et al. Genetic instability and mammary tumor formation in mice carrying mammary-specific disruption of Chk1 and p53. Oncogene 2010; 29:4007 - 17; http://dx.doi.org/10.1038/onc.2010.163; PMID: 20473325
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 2001; 409:355 - 9; http://dx.doi.org/10.1038/35053094; PMID: 11201745
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 2000; 101:635 - 45; http://dx.doi.org/10.1016/S0092-8674(00)80875-2; PMID: 10892650
- Garber PM, Rine J. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 2002; 161:521 - 34; PMID: 12072451
- Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol 2002; 12:1797 - 806; http://dx.doi.org/10.1016/S0960-9822(02)01226-5; PMID: 12419179
- Cheung HW, Jin DY, Ling MT, Wong YC, Wang Q, Tsao SW, et al. Mitotic arrest deficient 2 expression induces chemosensitization to a DNA-damaging agent, cisplatin, in nasopharyngeal carcinoma cells. Cancer Res 2005; 65:1450 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-04-0567; PMID: 15735033
- Fung MK, Han HY, Leung SC, Cheung HW, Cheung AL, Wong YC, et al. MAD2 interacts with DNA repair proteins and negatively regulates DNA damage repair. J Mol Biol 2008; 381:24 - 34; http://dx.doi.org/10.1016/j.jmb.2008.05.080; PMID: 18597777
- Carrassa L, Chilà R, Lupi M, Ricci F, Celenza C, Mazzoletti M, et al. Combined inhibition of Chk1 and Wee1: in vitro synergistic effect translates to tumor growth inhibition in vivo. Cell Cycle 2012; 11:2507 - 17; http://dx.doi.org/10.4161/cc.20899; PMID: 22713237
- Leung-Pineda V, Ryan CE, Piwnica-Worms H. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol Cell Biol 2006; 26:7529 - 38; http://dx.doi.org/10.1128/MCB.00447-06; PMID: 17015476
- Wassmann K, Liberal V, Benezra R. Mad2 phosphorylation regulates its association with Mad1 and the APC/C. EMBO J 2003; 22:797 - 806; http://dx.doi.org/10.1093/emboj/cdg071; PMID: 12574116
- O’Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, et al. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem 2000; 275:22719 - 27; http://dx.doi.org/10.1074/jbc.M001002200; PMID: 10801797
- Hutchins JR, Hughes M, Clarke PR. Substrate specificity determinants of the checkpoint protein kinase Chk1. FEBS Lett 2000; 466:91 - 5; http://dx.doi.org/10.1016/S0014-5793(99)01763-9; PMID: 10648819
- Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, et al. The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell 2000; 100:681 - 92; http://dx.doi.org/10.1016/S0092-8674(00)80704-7; PMID: 10761933
- Liu Q, Hirohashi Y, Du X, Greene MI, Wang Q. Nek2 targets the mitotic checkpoint proteins Mad2 and Cdc20: a mechanism for aneuploidy in cancer. Exp Mol Pathol 2010; 88:225 - 33; http://dx.doi.org/10.1016/j.yexmp.2009.12.004; PMID: 20034488
- Wang J, Han X, Feng X, Wang Z, Zhang Y. Coupling cellular localization and function of checkpoint kinase 1 (Chk1) in checkpoints and cell viability. J Biol Chem 2012; 287:25501 - 9; http://dx.doi.org/10.1074/jbc.M112.350397; PMID: 22692200
- Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, Roopra A, et al. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci USA 2012; 109:E2205 - 14; http://dx.doi.org/10.1073/pnas.1201911109; PMID: 22778409
- Enomoto M, Goto H, Tomono Y, Kasahara K, Tsujimura K, Kiyono T, et al. Novel positive feedback loop between Cdk1 and Chk1 in the nucleus during G2/M transition. J Biol Chem 2009; 284:34223 - 30; http://dx.doi.org/10.1074/jbc.C109.051540; PMID: 19837665