Abstract
Caveolin-1 (Cav-1) is a critical regulator of tumor progression in a variety of cancers where it has been shown to act as either a tumor suppressor or tumor promoter. In glioblastoma multiforme, it has been previously demonstrated to function as a putative tumor suppressor. Our studies here, using the human glioblastoma-derived cell line U-87MG, further support the role of Cav-1 as a negative regulator of tumor growth. Using a lentiviral transduction approach, we were able to stably overexpress Cav-1 in U-87MG cells. Gene expression microarray analyses demonstrated significant enrichment in gene signatures corresponding to downregulation of MAPK, PI3K/AKT and mTOR signaling, as well as activation of apoptotic pathways in Cav-1-overexpressing U-87MG cells. These same gene signatures were later confirmed at the protein level in vitro. To explore the ability of Cav-1 to regulate tumor growth in vivo, we further show that Cav-1-overexpressing U-87MG cells display reduced tumorigenicity in an ectopic xenograft mouse model, with marked hypoactivation of MAPK and PI3K/mTOR pathways. Finally, we demonstrate that Cav-1 overexpression confers sensitivity to the most commonly used chemotherapy for glioblastoma, temozolomide. In conclusion, Cav-1 negatively regulates key cell growth and survival pathways and may be an effective biomarker for predicting response to chemotherapy in glioblastoma.
Glioblastoma multiforme (GBM) is the most common and most deadly primary brain tumor affecting adults. Despite advancements made in surgical, radiological and chemo-therapies for this grade IV astrocytoma, prognoses have remained very poor: median survival time from diagnosis remains at 9–15 mo, with less than 10% of patients surviving beyond 5 y.Citation1,Citation2
Caveolin-1 (Cav-1) is the principle structural protein responsible for the formation of caveolae, or invaginating microdomains, in the cell membrane. The capacity for Cav-1 to associate with a wide variety of proteins has implicated it in a number of processes, ranging from vesicular transport and cholesterol homeostasis to nitric oxide production and cell migration, among others.Citation3-Citation7 Its ability to regulate cell cycle progression and intracellular signal transduction have resulted in the substantial characterization of Cav-1 in many cancers, where it has been shown to act as both a tumor suppressor and tumor promoter depending on the tissue type.Citation8-Citation11 In gliomas, expression of Cav-1 appears to increase proportionally to tumor grade, with most GBM lesions exhibiting more intense Cav-1 immunoreactivity than their grade II and III counterparts.Citation12-Citation14 However, little is currently known as to the role of Cav-1 as it relates to GBM in vivo. Recent in vitro studies conducted using the GBM-derived cell line U-87MG have demonstrated that Cav-1 acts as a putative tumor suppressor in GBM by downregulating α5β1 integrin expression and subsequent TGFβ/SMAD pathway activity.Citation15,Citation16 Consistent with these findings, we here show that U-87MG cells stably overexpressing Cav-1 exhibit diminished mitogenic signaling, upregulated activation of apoptotic pathways and a significantly decreased ability to form tumors in vivo. Additionally, we show that expression of Cav-1 confers sensitivity to the alkylating agent temozolomide (TMZ), the most commonly used chemotherapy for GBM. These studies further support the role of Cav-1 as a putative tumor suppressor in gliomas and serve to underscore the potential of Cav-1 to serve as a favorable prognostic factor in GBM.
Results
Stable expression of Cav-1 in U-87MG cells
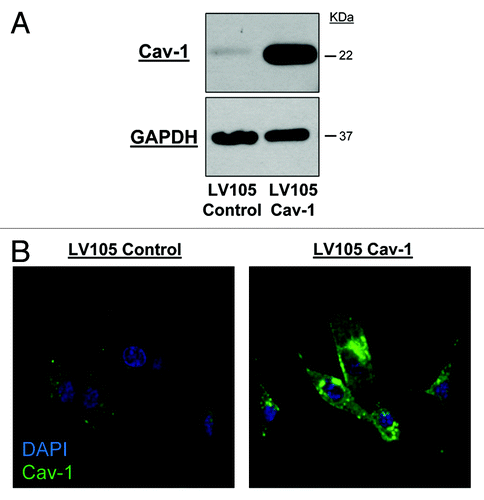
In order to establish durable expression of Cav-1 over time in a cell line model, we chose to use a lentiviral transduction approach over the transient transfection methods used in previous in vitro studies.Citation15,Citation16 After selection with puromycin, U-87MG cells transduced with lentiviral constructs stably expressing full length Cav-1 cDNA (LV105 Cav-1) were shown to effectively upregulate Cav-1 compared with an empty control lentivirus (LV105 Control) as demonstrated by western immunoblot (). Changes in Cav-1 protein expression were also confirmed by immunofluorescence, where overexpressing cells demonstrated increased cytoplasmic and membrane localization of Cav-1 following lentiviral transduction ().
Cav-1 regulates cancer-associated gene expression
Using a microarray consisting of > 20,000 transcript probes, we were able to identify 2,001 genes (~10%) significantly modulated by Cav-1 overexpression ( and ; Tables S1, S2 and S3). Gene set enrichment analyses performed on microarray expression data obtained from LV105 control and LV105 Cav-1 U-87MG cells indicates that Cav-1 expression corresponds to changes in a variety of cancer-associated gene signatures. Specifically, by comparing expression data to biological process gene ontology sets, it was found that Cav-1-overexpressing U-87MG cells demonstrated significant (p < 0.001) enrichment among gene sets related to negative regulation of signal transduction, MAP-kinase activity, cell proliferation and transcription (; Table S1). Signatures related to caspase activation, apoptosis and the transforming growth factor β pathway were also highly enriched (; Table S1). When expression data was compared with a curated canonical pathway database, gene sets related to PI3K/AKT, mTOR and ERK signaling, as well as cell death and extracellular matrix signaling, were found to be significantly enriched (; Table S1).
Table 1: Differential gene expression of Cav-1 overexpressing U-87MG cells
Table 2. Cav-1 regulates cancer-associated gene expression
Cav-1 mediates major proliferative and cell-survival pathways
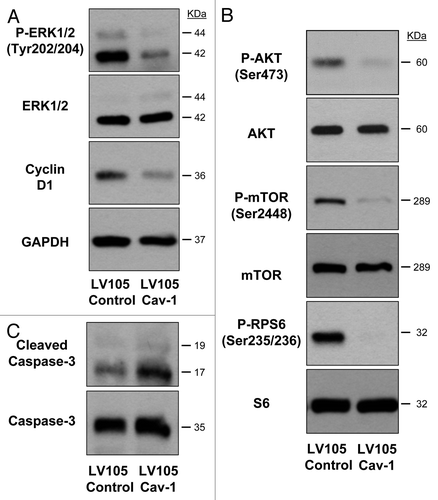
To validate the results obtained from our microarray analyses, we next sought to confirm Cav-1 mediated modulation of intracellular signaling pathways at the protein level. Overexpression of Cav-1 in U-87MG cells results in abrogated activity of proliferative pathways as shown by reduced phosphorylation of ERK1/2 and decreased expression of the cell cycle driver cyclin D1 when compared with control as shown by western immunoblot (). Overexpression of Cav-1 further reduced the activity of protein synthesis pathways as shown by reduced activation of the AKT/mTOR/ribosomal protein S6 (RPS6) signaling pathways (). Additionally, U-87MG cells overexpressing Cav-1 demonstrated increased presence of the apoptosis activator cleaved caspase 3 ().
Figure 2. Cav-1 mediates major proliferative and cell survival pathways. Western immunoblot analysis of LV105 control and LV105 Cav-1 U-87MG cells showing cyclin D1 and cleaved caspase-3 expression as well as phosphorylation status of ERK1/2, AKT, mTOR and RPS6 pathways, with respective total protein levels for loading controls. Total caspase-3 and GAPDH serve as loading controls for cleaved caspase-3 and cyclin D1, respectively.
U-87MG cells stably overexpressing Cav-1 exhibit decreased tumor growth in vivo
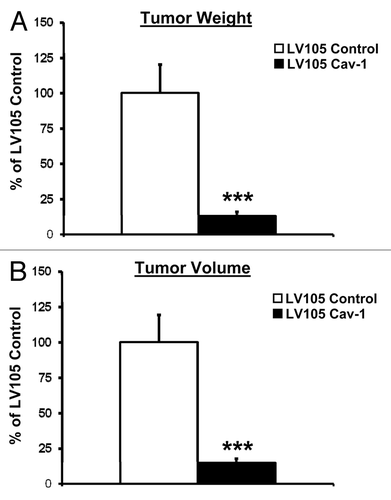
To evaluate the ability of Cav-1 to regulate tumorigenicity in vivo, U-87MG cells infected with either Cav-1-expressing or control lentivirus were injected subcutaneously into the flanks of athymic nu/nu male mice. After 4 wk, mice were sacrificed and tumors were collected, weighed and measured. Importantly, mice harboring Cav-1 overexpressing tumors demonstrated markedly reduced (~7-fold) tumor weights and volumes as compared with their control counterparts (p < 0.001, ).
Cav-1 overexpressing tumors show reduced signaling activity in vivo
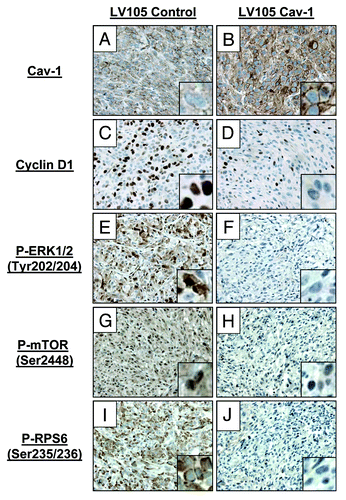
Similar to results obtained in vitro, our immunohistochemical analyses show that explanted xenograft tumors overexpressing Cav-1 demonstrate fewer cells staining positive for phospho-ERK1/2 and cyclin D1 (). Additionally, RPS6 and MTOR pathways were shown to be silenced in LV105 Cav-1 tumors, as shown by the absence of their active, phosphorylated isoforms ().
Cav-1 confers chemosensitivity in U-87MG cells
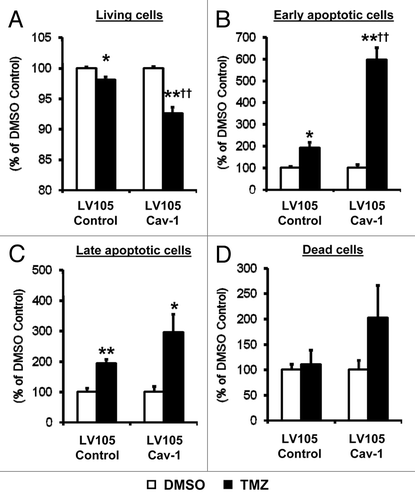
To further examine the effect of Cav-1 on chemotherapeutic-induced apoptosis, U-87MG cells were stained for the cell death marker Annexin V and measured by flow cytometry. Cav-1-overexpressing U-87MG cells cultured for 72 h in the presence of 500 μM of temozolomide (TMZ), the most commonly used chemotherapeutic for GBM, showed significant reductions in cell viability when compared with TMZ-treated LV105 control cells (5.5%, p < 0.01, ). This effect was found to be most pronounced among cells initiating apoptosis, as Cav-1-overexpressing U-87MG cells treated with TMZ demonstrated > 400% increase in early apoptotic cells compared with TMZ-treated LV105 control cells (p < 0.01, ). Interestingly, although not statistically significant, overexpression of Cav-1 yielded expanded late-apoptotic and dead cells after 72 h of TMZ treatment ().
Figure 5. Cav-1 confers chemosensitivity in U-87MG cells. Annexin V and propidium iodide staining of U-87MG cells treated with either DMSO control or 500 μm TMZ reveals percentages of (A) live cells, (B) early apoptotic cells, (C) late apoptotic cells and (D) dead cells as measured by flow cytometry. Each group is normalized to its own DMSO control (*p < 0.05 and **p < 0.01 vs. internal DMSO control, †p < 0.01 vs. TMZ treated LV105 control, n = 3 per group).
Discussion
Although it has been demonstrated that Cav-1 expression in glioma increases variably in accordance with grade, little is currently known about its biological effects on tumor onset and progression.Citation12-Citation14,Citation17-Citation19 Previous in vitro studies using transient transfection techniques have shown that loss of Cav-1 in U-87MG cells resulted in the adoption of a more proliferative and invasive phenotype, whereas its forced overexpression conferred the opposite effects.Citation15 In line with previous studies, we here show that Cav-1 functions as a putative tumor suppressor in glioblastoma. Using a novel lentivirus transduction system we created a stable Cav-1 overexpressing cell line based on the U-87MG background.
By subjecting transiently transfected U-87MG cells to a panel of reverse transcription-PCR primers, Martin et al. identified genes pertaining to cell invasion, metastasis and cell adhesion as those being the most differentially regulated by Cav-1 expression. Particularly, they showed that the integrin genes ITGA1, ITGA3, ITGA5, ITGAV, ITGB1 and ITGB5 were significantly downregulated in Cav-1-overexpressing cells, with cells treated with Cav-1-specific siRNA demonstrating marked upregulation of these same genes. Matrix metallopeptidase genes MMP1 and MMP2 as well as transforming growth factor β receptor I (TGFRBI) were also shown to be significantly modulated by Cav-1.Citation15 A follow-up study using Cav-1-silenced U-87MG cells further clarified a mechanism in which Cav-1 acts as a negative regulator of integrin signaling by inhibiting the expression of these integrins themselves as well as sequestering downstream TGFβ/TGFβRI/SMAD2 and ERK pathways.Citation16 Here, we implemented a similar, albeit much more expansive, microarray-based approach to study gene perturbations as a result of Cav-1 overexpression. Using gene set enrichment analyses, we indeed show similar expression profiles to those found previously, with gene sets related to integrin interactions, as well as regulation of TGFβ receptor/SMAD pathways showing significant enrichment. In our study, however, we detected a multitude of other significantly enriched gene sets that have not been demonstrated previously in Cav-1-expressing GBM cells. For instance, U-87MG cells overexpressing Cav-1 demonstrated significant upregulation of genes responsible for negative regulation of signal transduction, particularly within ERK, PI3K/AKT and mTOR pathways. Although it has long been known that Cav-1 serves to negatively regulate the activity of p42/p44 (ERK1/2) signaling proteins of the MAPK pathway, our evidence also suggests it has the capability to sequester PI3K and mTOR activity.Citation20-Citation26 This is notable due to the fact that ERK, PI3K and mTOR signaling axes are frequently upregulated in GBM, suggesting that loss of Cav-1 could lead to unchecked activation of these pathways.Citation27-Citation31 Two of the most commonly silenced genes in GBM are the tumor suppressor proteins PTEN and TP53, which serve to antagonize the PI3K/AKT/mTOR pathway and regulate cell cycle response to DNA damage and cell death, respectively.Citation32 Of note is that these two genes were among the most upregulated in cells overexpressing Cav-1, which would likely explain the gene signatures corresponding to downregulation of signaling pathways and reduced invasiveness.
A major hallmark of GBM is the ability of tumor cells to invariably metastasize to distant sites in the CNS despite aggressive treatment. This is often attributed to the excessive release of matrix metallopeptidases and urokinase plasminogen activator.Citation33 Here we show that the genes MMP1, MMP3 and PLAU (urokinase plasminogen activator) are highly downregulated in our Cav-1-overexpressing U-87MG cells, which is consistent with reports that Cav-1 negatively regulates tumor invasiveness.Citation15,Citation34-Citation36 These genes have been shown to be regulated by Erk and TP53, therefore, their reduction may be secondary to Cav-1 modulation of these pathways.Citation37-Citation40 Of note, we also found that genes responsible for sequestering cell cycle progression and transcription were overexpressed in LV105 Cav-1 cells (FOXN3, HDAC5, VHL, CDKN1C, among others). Conversely, genes responsible for progression through cell cycle, such as CCND1 (cyclin D1), were found to be significantly downregulated in Cav-1-overexpressing cells, consistent with previous reports that Cav-1 transcriptionally represses cyclin D1.Citation8 Perhaps our most notable finding, however, is that a substantial number of genes involved in the activation of apoptotic and cell death pathways are increased as a result of Cav-1 overexpression (TP53, MOAP1, CASP3, CASP9, BCL2L11, BAK1, BID among others). Although the role of Cav-1 in apoptosis is contentious, with reports indicating both pro- and anti-cell death roles, it may be possible that expression of Cav-1 promotes apoptotic activity in U-87MG cells by inhibiting the BIRC5 gene product, survivin, as is suggested here and in previous reports.Citation41-Citation45 In support of these microarray data, we were able to demonstrate, at the protein level, silencing of ERK, AKT, mTOR, RPS6 and cyclin D1 pathways with corresponding activation and cleavage of the key apoptosis initiator caspase 3.
Importantly, we here show for the first time that forced expression of Cav-1 in vivo results in a dramatic reduction of tumor burden in U-87MG xenografts. Although Cosset et al. have demonstrated that explanted human glioma tissue lacking Cav-1 expression results in increased expression of α5β1 integrin subunits, we were able to demonstrate a direct inverse relationship with Cav-1 expression and cell proliferation in an animal model.Citation16 In line with our in vitro data, these xenograft tumors displayed reduced activity of ERK, RPS6 and mTOR pathways. As these pathways have been previously shown to play major roles in glioma progression, it is likely that Cav-1 could act as a critical regulator of tumor growth and protein synthesis in a clinical setting. As examples of this, studies have shown that exogenous administration of cavtratin, or a soluble peptide consisting of the Cav-1 scaffolding domain fused to an internalization domain, results in reduced MAPK activity in oligodendroglial cells in vivo, as well as reduced tumor volumes in a xenograft model of Lewis lung carcinoma.Citation23,Citation46 A separate study demonstrated that in vitro administration of full-length Cav-1 prevented invasion of three different GBM-derived cell lines using a Boyden-chamber assay.Citation47 In this regard, it may be suggested that Cav-1 be explored as a therapeutic agent in GBM.
Lastly, we have also demonstrated that expression of Cav-1 confers sensitivity to the most commonly used chemotherapeutic in GBM, temozolomide. This could be due in part to the action of the permeability glycoprotein (P-gp) transporter, a multidrug exporter that normally prevents the influx of drugs across the blood brain barrier. However, cancerous cells can also express this protein, rendering treatment of GBM with conventional chemotherapeutics less effective.Citation48 Cav-1 has been shown to associate with P-gp and negatively regulate its activity; therefore, overexpression of Cav-1 most likely results in improved access of TMZ to the intracellular compartment of U-87MG cells in our model.Citation49,Citation50 Interestingly, a separate study showed that treatment with TMZ resulted in upregulation of Cav-1 expression in vivo using orthotopic GBM xenograft models.Citation47 In light of our data, this could suggest a positive feedback loop exists in which treatment with TMZ serves to auto-sensitize GBM cells through a Cav-1 dependent mechanism. This finding implicates Cav-1 as a potential biomarker predicting response to chemotherapies for GBM, as it has been shown for other cancers such as breast, lung and oral squamous cell carcinomas.Citation51-Citation53
Taken together, these studies confirm and expand upon previous work identifying Cav-1 as a putative tumor suppressor in GBM. We here show that stable overexpression of Cav-1 in a widely used model of GBM results in silencing of key proliferative and cell survival pathways in vitro as well as in vivo (). Additionally, we have demonstrated its ability to modulate sensitivity to commonplace chemotherapeutics for GBM. These findings highlight the potential of Cav-1 to serve as a novel biomarker indicating potential response to therapy and also a candidate therapy for treatment of GBM.
Materials and Methods
Cell lines and reagents
The human glioblastoma-derived cell line U-87MG was obtained from American Type Culture Collection (ATCC) and cultured in Eagle’s modified essential medium (EMEM, ATCC) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Life Technologies). Cells were cultured in the presence of 5% CO2 at 37°C. Temozolomide (TMZ) was obtained from Sigma-Aldrich and dissolved in DMSO to a concentration of 100 mM. The following antibodies were used: mouse anti-Caveolin-1 (2297, BD Bioscience), rabbit anti-Caveolin-1 (N-20, Santa Cruz Biotechnology), mouse anti-cyclin D1 (DCS-6, Santa Cruz Biotechnology), rabbit anti-phospho mTOR (Se2448, D9C2, Cell Signaling), rabbit anti-phospho-ERK1/2 (Thr202/Tyr204, Cell Signaling), rabbit anti-phospho-AKT (Ser473, D9E, Cell Signaling), rabbit anti-phospho-ribosomal S6 (Ser235/236, 91B2, Cell Signaling) rabbit anti-ERK1/2 (Cell Signaling), rabbit anti-AKT (Cell Signaling), rabbit anti-ribosomal S6 (5G10, Cell Signaling), rabbit anti-cleaved caspase 3 (Asp175, Cell Signaling), rabbit anti-caspase 3 (Cell Signaling) and mouse anti-GAPDH (6C5, Fitzgerald Industries).
Stable lentiviral transduction of U-87MG cells
Plasmids Ex-Neg-LV105 (empty control vector) and Ex-D0159-LV105 (Cav-1 cDNA vector) were obtained from Genecopoeia and transfected into the packaging cell line Genecopoeia 293Ta using the Lenti-Pac HIV Expression Packaging Kit as per manufacturer’s instructions. Forty-eight h post-transfection, lentivirus containing supernatants were collected and centrifuged at 500 × g for 10 min to clear cellular debris. U-87MG cells were cultured in viral supernatants supplemented with 5 μg/ml polybrene (Santa Cruz) for 24 h prior to changing back into complete medium containing 2.5 μg/ml puromycin hydrochloride (Santa Cruz) to select for lentiviral-transduced cells. After 1 wk of selection, cells were allowed to grow in complete medium without puromycin.
Western immunoblot
Cells at 70% confluence were collected, pelleted at 300 × g, washed twice with Dulbecco’s PBS (DPBS) and resuspended in RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, pH 7.5) including Complete Protease Inhibitor Cocktail (Roche Diagnostics) and Halt Phosphatase Inhibitor Cocktail (Thermo-Scientific). Lysates were sonicated and centrifuged at 10,000 × g for 10 min to clear cellular debris prior to protein quantification by BCA assay (Thermo-Scientific) as per manufacturer’s instructions. Proteins were separated by sodium dodecyl sulfate PAGE (SDS-PAGE; 8–12% acrylamide), transferred to nitrocellulose membranes (Whatman) and blocked for 1 h in TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween-20, pH 8.0) with 5% bovine serum albumin (BSA). Membranes were incubated with primary antibodies diluted in TBST + 1% BSA overnight at 4°C followed by incubation in either horseradish peroxidase (HRP) conjugated anti-mouse (Thermo-Scientific) or anti-rabbit (BD Biosciences) antibodies. Detection of bound antibodies was accomplished with the use of Supersignal chemiluminescent substrates (Thermo-Scientific).
Immunofluorescence analysis
U-87MG cells grown on glass coverslips in 6-well plates were fixed in ice-cold methanol for 20 min, washed with PBS and incubated with anti-Cav-1 primary antibody (BD Bioscience) in immunofluorescence (IF) buffer (PBS + 5% BSA, 0.5% NP40) for 30 min at 37°C before incubation with secondary fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody (Jackson Labs) in IF buffer. Cells were counterstained with Hoechst nuclear dye (Life Technologies) prior to coverslipping and visualization with a Zeiss LSM 510 confocal microscope (Carl Zeiss Microscopy).
Gene array
DNA microarray analysis was performed using the Human Whole Genome OneArray v2 (Phalanx Biotech). RNA quality and integrity were determined utilizing an Agilent 2100 Bioanalyzer (Agilent Technologies) and absorbance at A260/A280. Only high quality RNA, having a RIN of > 7.0 and an A260/280 absorbance ratio of > 1.8, was utilized for further experimentation. RNA was synthesized to double-stranded cDNA and amplified using in vitro transcription that included amino-allyl UTP, and the aRNA product was subsequently conjugated with Cy5 NHS ester (GE Healthcare Lifesciences). Fragmented aRNA was hybridized at 42°C overnight using the HybBag mixing system with 1× OneArray Hybridization Buffer (Phalanx Biotech), 0.01 mg/ml sheared salmon sperm DNA (Promega), at a concentration of 0.025 mg/ml labeled target. After hybridization, the arrays were washed according to the OneArray protocol. Raw intensity signals for each microarray were captured using a Molecular Dynamics™ Axon 4100A scanner, measured using GenePixPro™ Software. Significantly up- or downregulated genes in LV105 Cav-1 cells were identified as having normalized intensities above background (> 50), a fold change of ± 1.5 compared with control and p < 0.05.
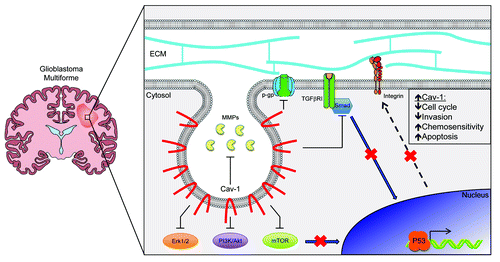
Figure 6. Schematic representation of the role of Cav-1 in glioblastoma. Gliomas are highly heterogeneous tumors that have been demonstrated to contain populations of cells with varied levels of Cav-1 expression. In this case, enhanced Cav-1 expression among a tumor cell prevents the activation of the TGFBRI/SMAD pathway, which, in turn, suppresses expression of integrin subunits at the transcriptional level and their subsequent signaling activity. Increased Cav-1 expression is also correlated with a decrease in the availability of matrix metallopeptidases, downregulated activity of the Erk1/2, PI3K/Akt and mTOR signaling pathways as well as the inhibition of the trans-membrane drug exporter p-gp. Together, this may implicate that tumors with increased Cav-1 levels are less likely to progress through the cell cycle or invade into surrounding tissues and are primed to undergo P53 mediated apoptosis, making these cells more easily targeted by standard chemotherapy regimens.
Gene set enrichment analyses
Pre-processed expression data was subjected to Gene Set Enrichment Analysis using C5.BP.V3.0 (gene ontology: biological processes) and C2.CP.V3.0 (canonical pathways) MSigDB gene sets.Citation54,Citation55 Genes expression data were ordered based on a Signal2Noise metric and compared statistically to existing gene sets at a resolution of 1,000 permutations. Statistical significance of gene set enrichment was assumed at nominal p < 0.05, with a false discovery rate (FDR) q < 0.25.
Tumor xenografts
All animal studies were conducted in accordance with the guidelines set forth by the National Institutes of Health and the Thomas Jefferson University Institutional Animal Care and Use Committee (IACUC). Briefly, U-87MG cells were washed with DPBS, trypsinized, counted and resuspended in a volume of complete medium yielding 1 × 106 cells/50 μl, which was subsequently injected subcutaneously into the flanks of 6–8-wk-old male athymic nu/nu mice (NCI). After 4 wk, mice were sacrificed and tumors were excised, weighed and measured prior to further processing.
Immunohistochemistry
Explanted xenograft tumors were fixed in 10% phosphate buffered formalin solution for 24 h prior to dehydration in 70% ethanol, paraffin embedding and sectioning onto slides. Following xylene deparaffinizaiton and rehydration, slides were subjected to 10 min of heat antigen retrieval in 10 mM sodium citrate buffer pH 6.0 and endogenous peroxide quenching in 3% hydrogen peroxide for 20 min. Tissues were blocked in 10% normal goat serum (NGS, Vector Labs) for 1 h at room temperature and incubated overnight with primary antibody in 10% NGS at 4°C. Slides were then washed in PBS and blocked with Biotin-Blocking System (Dako) before incubating with the appropriate secondary antibody in PBS and developing with 3,3-diaminobenzidine (DAB) substrate (Dako). Slides were counterstained with hematoxylin (Sigma), dehydrated and coverslipped prior to imaging with an Olympus BX51 light microscope equipped with a Micropublisher 5.0 CCD camera (QImaging).
Flow cytometry
U-87MG cells (50,000/well) were plated in 12-well tissue culture dishes and allowed to attach overnight prior to changing their medium into complete EMEM containing either 500 μM TMZ or DMSO control and culturing them for an additional 72 h. Cells were then trypsinized, centrifuged at 300 × g for 5 min and resuspended in binding buffer with APC-conjugated anti-Annexin V antibody (BD Biosciences) and 0.33 μg/ml Propidium Iodide (PI, KPL). All samples were run on a BD FACSCalibur flow cytometer (BD Biosciences). Cells were quantified according to staining as follows: viable (Annexin V-negative, PI-negative), early apoptotic (Annexin V-positive, PI-negative), late apoptotic (Annexin V-positive, PI-positive) and dead (Annexin V-negative, PI-positive).
Statistical analysis
All data were expressed as mean ± SEM. Differences between groups were evaluated by either unpaired Student’s t-test or one-way ANOVA followed by Tukey’s multiple-group comparisons test, where appropriate. Statistical significance was assumed at p < 0.05.
| Abbreviations: | ||
| Cav-1 | = | Caveolin-1 |
| GBM | = | glioblastoma multiforme |
| MAPK | = | mitogen activated protein kinase |
| PI3K | = | phosphatidylinositol 3-kinase |
| RPS6 | = | ribosomal protein S6 |
| mTOR | = | mammalian target of rapamycin |
| TMZ | = | temozolomide |
| TGFβ | = | transforming growth factor beta |
| TGFβRI | = | transforming growth factor beta receptor I |
| TP53 | = | tumor protein 53 |
| PTEN | = | phosphatase and tensin homolog |
| p-gp | = | permeability glycoprotein |
Additional material
Download Zip (10.2 MB)Acknowledgments
Jean-Francois Jasmin was supported by a Susan G. Komen Career Catalyst Research Grant. Michael P. Lisanti and his laboratory were supported via the resources of Thomas Jefferson University.
Disclosure of Ptoential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/24497
References
- Buckner JC, Brown PD, O’Neill BP, Meyer FB, Wetmore CJ, Uhm JH. Central nervous system tumors. Mayo Clin Proc 2007; 82:1271 - 86; http://dx.doi.org/10.4065/82.10.1271; PMID: 17908533
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al, European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups, National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009; 10:459 - 66; http://dx.doi.org/10.1016/S1470-2045(09)70025-7; PMID: 19269895
- van Deurs B, Roepstorff K, Hommelgaard AM, Sandvig K. Caveolae: anchored, multifunctional platforms in the lipid ocean. Trends Cell Biol 2003; 13:92 - 100; http://dx.doi.org/10.1016/S0962-8924(02)00039-9; PMID: 12559760
- García-Cardeña G, Martasek P, Masters BS, Skidd PM, Couet J, Li S, et al. Dissecting the interaction between nitric oxide synthase (NOS) and caveolin. Functional significance of the nos caveolin binding domain in vivo. J Biol Chem 1997; 272:25437 - 40; http://dx.doi.org/10.1074/jbc.272.41.25437; PMID: 9325253
- Navarro A, Anand-Apte B, Parat MO. A role for caveolae in cell migration. FASEB J 2004; 18:1801 - 11; http://dx.doi.org/10.1096/fj.04-2516rev; PMID: 15576483
- Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol 2007; 8:185 - 94; http://dx.doi.org/10.1038/nrm2122; PMID: 17318224
- Razani B, Woodman SE, Lisanti MP. Caveolae: from cell biology to animal physiology. Pharmacol Rev 2002; 54:431 - 67; http://dx.doi.org/10.1124/pr.54.3.431; PMID: 12223531
- Hulit J, Bash T, Fu M, Galbiati F, Albanese C, Sage DR, et al. The cyclin D1 gene is transcriptionally repressed by caveolin-1. J Biol Chem 2000; 275:21203 - 9; http://dx.doi.org/10.1074/jbc.M000321200; PMID: 10747899
- Patel HH, Murray F, Insel PA. Caveolae as organizers of pharmacologically relevant signal transduction molecules. Annu Rev Pharmacol Toxicol 2008; 48:359 - 91; http://dx.doi.org/10.1146/annurev.pharmtox.48.121506.124841; PMID: 17914930
- Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol 2005; 288:C494 - 506; http://dx.doi.org/10.1152/ajpcell.00458.2004; PMID: 15692148
- Goetz JG, Lajoie P, Wiseman SM, Nabi IR. Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metastasis Rev 2008; 27:715 - 35; http://dx.doi.org/10.1007/s10555-008-9160-9; PMID: 18506396
- Cassoni P, Senetta R, Castellano I, Ortolan E, Bosco M, Magnani I, et al. Caveolin-1 expression is variably displayed in astroglial-derived tumors and absent in oligodendrogliomas: concrete premises for a new reliable diagnostic marker in gliomas. Am J Surg Pathol 2007; 31:760 - 9; http://dx.doi.org/10.1097/01.pas.0000213433.14740.5d; PMID: 17460461
- Barresi V, Buttarelli FR, Vitarelli EE, Arcella A, Antonelli M, Giangaspero F. Caveolin-1 expression in diffuse gliomas: correlation with the proliferation index, epidermal growth factor receptor, p53, and 1p/19q status. Hum Pathol 2009; 40:1738 - 46; http://dx.doi.org/10.1016/j.humpath.2009.04.026; PMID: 19716156
- Senetta R, Trevisan E, Rudà R, Maldi E, Molinaro L, Lefranc F, et al. Caveolin 1 expression independently predicts shorter survival in oligodendrogliomas. J Neuropathol Exp Neurol 2009; 68:425 - 31; http://dx.doi.org/10.1097/NEN.0b013e31819ed0b7; PMID: 19287309
- Martin S, Cosset EC, Terrand J, Maglott A, Takeda K, Dontenwill M. Caveolin-1 regulates glioblastoma aggressiveness through the control of alpha(5)beta(1) integrin expression and modulates glioblastoma responsiveness to SJ749, an alpha(5)beta(1) integrin antagonist. Biochim Biophys Acta 2009; 1793:354 - 67; http://dx.doi.org/10.1016/j.bbamcr.2008.09.019; PMID: 18992284
- Cosset EC, Godet J, Entz-Werlé N, Guérin E, Guenot D, Froelich S, et al. Involvement of the TGFβ pathway in the regulation of α5 β1 integrins by caveolin-1 in human glioblastoma. Int J Cancer 2012; 131:601 - 11; http://dx.doi.org/10.1002/ijc.26415; PMID: 21901744
- Abulrob A, Giuseppin S, Andrade MF, McDermid A, Moreno M, Stanimirovic D. Interactions of EGFR and caveolin-1 in human glioblastoma cells: evidence that tyrosine phosphorylation regulates EGFR association with caveolae. Oncogene 2004; 23:6967 - 79; http://dx.doi.org/10.1038/sj.onc.1207911; PMID: 15273741
- Forget MA, Desrosiers RR, Del M, Moumdjian R, Shedid D, Berthelet F, et al. The expression of rho proteins decreases with human brain tumor progression: potential tumor markers. Clin Exp Metastasis 2002; 19:9 - 15; http://dx.doi.org/10.1023/A:1013884426692; PMID: 11918088
- Cameron PL, Liu C, Smart DK, Hantus ST, Fick JR, Cameron RS. Caveolin-1 expression is maintained in rat and human astroglioma cell lines. Glia 2002; 37:275 - 90; http://dx.doi.org/10.1002/glia.10036; PMID: 11857686
- Engelman JA, Chu C, Lin A, Jo H, Ikezu T, Okamoto T, et al. Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Lett 1998; 428:205 - 11; http://dx.doi.org/10.1016/S0014-5793(98)00470-0; PMID: 9654135
- Cohen AW, Park DS, Woodman SE, Williams TM, Chandra M, Shirani J, et al. Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. Am J Physiol Cell Physiol 2003; 284:C457 - 74; PMID: 12388077
- Han F, Gu D, Chen Q, Zhu H. Caveolin-1 acts as a tumor suppressor by down-regulating epidermal growth factor receptor-mitogen-activated protein kinase signaling pathway in pancreatic carcinoma cell lines. Pancreas 2009; 38:766 - 74; http://dx.doi.org/10.1097/MPA.0b013e3181b2bd11; PMID: 19893453
- Schmitz M, Zerr I, Althaus HH. Effect of cavtratin, a caveolin-1 scaffolding domain peptide, on oligodendroglial signaling cascades. Cell Mol Neurobiol 2011; 31:991 - 7; http://dx.doi.org/10.1007/s10571-011-9694-1; PMID: 21523467
- Xia H, Khalil W, Kahm J, Jessurun J, Kleidon J, Henke CA. Pathologic caveolin-1 regulation of PTEN in idiopathic pulmonary fibrosis. Am J Pathol 2010; 176:2626 - 37; http://dx.doi.org/10.2353/ajpath.2010.091117; PMID: 20395445
- Feng S, Wang Y, Wang X, Wang Z, Cui Y, Liu J, et al. Caveolin-1 gene silencing promotes the activation of PI3K/AKT dependent on Eralpha36 and the transformation of MCF10ACE. Science China. Life Sci 2010; 53:598 - 605; http://dx.doi.org/10.1007/s11427-010-0100-x
- Mercier I, Camacho J, Titchen K, Gonzales DM, Quann K, Bryant KG, et al. Caveolin-1 and accelerated host aging in the breast tumor microenvironment: chemoprevention with rapamycin, an mTOR inhibitor and anti-aging drug. Am J Pathol 2012; 181:278 - 93; http://dx.doi.org/10.1016/j.ajpath.2012.03.017; PMID: 22698676
- Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer 2010; 10:319 - 31; http://dx.doi.org/10.1038/nrc2818; PMID: 20414201
- Kita D, Yonekawa Y, Weller M, Ohgaki H. PIK3CA alterations in primary (de novo) and secondary glioblastomas. Acta Neuropathol 2007; 113:295 - 302; http://dx.doi.org/10.1007/s00401-006-0186-1; PMID: 17235514
- Sunayama J, Matsuda K, Sato A, Tachibana K, Suzuki K, Narita Y, et al. Crosstalk between the PI3K/mTOR and MEK/ERK pathways involved in the maintenance of self-renewal and tumorigenicity of glioblastoma stem-like cells. Stem Cells 2010; 28:1930 - 9; http://dx.doi.org/10.1002/stem.521; PMID: 20857497
- Mizoguchi M, Betensky RA, Batchelor TT, Bernay DC, Louis DN, Nutt CL. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol 2006; 65:1181 - 8; http://dx.doi.org/10.1097/01.jnen.0000248549.14962.b2; PMID: 17146292
- Lopez-Gines C, Gil-Benso R, Benito R, Mata M, Pereda J, Sastre J, et al. The activation of ERK1/2 MAP kinases in glioblastoma pathobiology and its relationship with EGFR amplification. Neuropathology 2008; 28:507 - 15; http://dx.doi.org/10.1111/j.1440-1789.2008.00911.x; PMID: 18410277
- Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol 2007; 170:1445 - 53; http://dx.doi.org/10.2353/ajpath.2007.070011; PMID: 17456751
- Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol 2006; 1:97 - 117; http://dx.doi.org/10.1146/annurev.pathol.1.110304.100043; PMID: 18039109
- Williams TM, Medina F, Badano I, Hazan RB, Hutchinson J, Muller WJ, et al. Caveolin-1 gene disruption promotes mammary tumorigenesis and dramatically enhances lung metastasis in vivo. Role of Cav-1 in cell invasiveness and matrix metalloproteinase (MMP-2/9) secretion. J Biol Chem 2004; 279:51630 - 46; http://dx.doi.org/10.1074/jbc.M409214200; PMID: 15355971
- Kim HN, Chung HS. Caveolin-1 inhibits membrane-type 1 matrix metalloproteinase activity. BMB Rep 2008; 41:858 - 62; http://dx.doi.org/10.5483/BMBRep.2008.41.12.858; PMID: 19123976
- Han F, Zhu HG. Caveolin-1 regulating the invasion and expression of matrix metalloproteinase (MMPs) in pancreatic carcinoma cells. J Surg Res 2010; 159:443 - 50; http://dx.doi.org/10.1016/j.jss.2009.03.079; PMID: 20031158
- Reunanen N, Westermarck J, Häkkinen L, Holmström TH, Elo I, Eriksson JE, et al. Enhancement of fibroblast collagenase (matrix metalloproteinase-1) gene expression by ceramide is mediated by extracellular signal-regulated and stress-activated protein kinase pathways. J Biol Chem 1998; 273:5137 - 45; http://dx.doi.org/10.1074/jbc.273.9.5137; PMID: 9478967
- Kim S, Lee Y, Lee DH, Kim Y, Cho KH, Chung JH. Basal and UV-induced MMP-1 expression are inhibited by p53 in human dermal fibroblasts. Exp Dermatol 2008; 17:939 - 45; http://dx.doi.org/10.1111/j.1600-0625.2008.00729.x; PMID: 18557930
- Sun Y, Sun Y, Wenger L, Rutter JL, Brinckerhoff CE, Cheung HS. p53 down-regulates human matrix metalloproteinase-1 (Collagenase-1) gene expression. J Biol Chem 1999; 274:11535 - 40; http://dx.doi.org/10.1074/jbc.274.17.11535; PMID: 10206959
- Shetty P, Velusamy T, Bhandary YP, Shetty RS, Liu MC, Shetty S. Urokinase expression by tumor suppressor protein p53: a novel role in mRNA turnover. Am J Respir Cell Mol Biol 2008; 39:364 - 72; http://dx.doi.org/10.1165/rcmb.2007-0406OC; PMID: 18390474
- Park J, Bae E, Lee C, Yoon SS, Chae YS, Ahn KS, et al. RNA interference-directed caveolin-1 knockdown sensitizes SN12CPM6 cells to doxorubicin-induced apoptosis and reduces lung metastasis. Tumour Biol 2010; 31:643 - 50; http://dx.doi.org/10.1007/s13277-010-0081-1; PMID: 20820979
- Yang X, Xiong H, Guan ZZ, Okai I, Ye D, Song Y, et al. Higher expression of Caveolin-1 inhibits human small cell lung cancer (SCLC) apoptosis in vitro. Cancer Invest 2012; 30:453 - 62; http://dx.doi.org/10.3109/07357907.2012.675384; PMID: 22545709
- Zhang M, Lee SJ, An C, Xu JF, Joshi B, Nabi IR, et al. Caveolin-1 mediates Fas-BID signaling in hyperoxia-induced apoptosis. Free Radic Biol Med 2011; 50:1252 - 62; http://dx.doi.org/10.1016/j.freeradbiomed.2011.02.031; PMID: 21382479
- Torres VA, Tapia JC, Rodríguez DA, Párraga M, Lisboa P, Montoya M, et al. Caveolin-1 controls cell proliferation and cell death by suppressing expression of the inhibitor of apoptosis protein survivin. J Cell Sci 2006; 119:1812 - 23; http://dx.doi.org/10.1242/jcs.02894; PMID: 16608879
- Zhang M, Lin L, Lee SJ, Mo L, Cao J, Ifedigbo E, et al. Deletion of caveolin-1 protects hyperoxia-induced apoptosis via survivin-mediated pathways. Am J Physiol Lung Cell Mol Physiol 2009; 297:L945 - 53; http://dx.doi.org/10.1152/ajplung.00081.2009; PMID: 19767411
- Lin MI, Yu J, Murata T, Sessa WC. Caveolin-1-deficient mice have increased tumor microvascular permeability, angiogenesis, and growth. Cancer Res 2007; 67:2849 - 56; http://dx.doi.org/10.1158/0008-5472.CAN-06-4082; PMID: 17363608
- Bruyère C, Abeloos L, Lamoral-Theys D, Senetta R, Mathieu V, Le Mercier M, et al. Temozolomide modifies caveolin-1 expression in experimental malignant gliomas in vitro and in vivo. Transl Oncol 2011; 4:92 - 100; PMID: 21461172
- Deeken JF, Löscher W. The blood-brain barrier and cancer: transporters, treatment, and Trojan horses. Clin Cancer Res 2007; 13:1663 - 74; http://dx.doi.org/10.1158/1078-0432.CCR-06-2854; PMID: 17363519
- Jodoin J, Demeule M, Fenart L, Cecchelli R, Farmer S, Linton KJ, et al. P-glycoprotein in blood-brain barrier endothelial cells: interaction and oligomerization with caveolins. J Neurochem 2003; 87:1010 - 23; http://dx.doi.org/10.1046/j.1471-4159.2003.02081.x; PMID: 14622130
- Guo Z, Zhu J, Zhao L, Luo Q, Jin X. Expression and clinical significance of multidrug resistance proteins in brain tumors. Journal of experimental & clinical cancer research. CR (East Lansing, Mich) 2010; 29:122
- Mercier I, Casimiro MC, Wang C, Rosenberg AL, Quong J, Minkeu A, et al. Human breast cancer-associated fibroblasts (CAFs) show caveolin-1 downregulation and RB tumor suppressor functional inactivation: Implications for the response to hormonal therapy. Cancer Biol Ther 2008; 7:1212 - 25; http://dx.doi.org/10.4161/cbt.7.8.6220; PMID: 18458534
- Ho CC, Kuo SH, Huang PH, Huang HY, Yang CH, Yang PC. Caveolin-1 expression is significantly associated with drug resistance and poor prognosis in advanced non-small cell lung cancer patients treated with gemcitabine-based chemotherapy. Lung Cancer 2008; 59:105 - 10; http://dx.doi.org/10.1016/j.lungcan.2007.07.024; PMID: 17850918
- Nakatani K, Wada T, Nakamura M, Uzawa K, Tanzawa H, Fujita S. Expression of caveolin-1 and its correlation with cisplatin sensitivity in oral squamous cell carcinoma. J Cancer Res Clin Oncol 2005; 131:445 - 52; http://dx.doi.org/10.1007/s00432-004-0662-8; PMID: 15856296
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545 - 50; http://dx.doi.org/10.1073/pnas.0506580102; PMID: 16199517
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al, The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25:25 - 9; http://dx.doi.org/10.1038/75556; PMID: 10802651