The eukaryotic translation initiation factor eIF4E is a potent oncogene elevated in an estimated 30% of human cancers.Citation1,Citation2 Its elevation in mouse models and tissue culture is linked to tumorigenesis and transformation, respectively. eIF4E acts in mRNA export and translation of specific transcripts by binding the methyl 7 guanosine cap found on the 5′ end of mRNAs. These transcripts typically encode proteins involved in proliferation, survival, invasion and metastases.Citation3,Citation4 The eIF4E family is comprised of three members:Citation5 the most commonly studied is referred to as eIF4E1 or eIF4E (as above); the second member is eIF4E2, also known as 4E-HP; and finally, the third member eIF4E3, which, to date, has been the least studied.
New findings suggest that, unlike eIF4E1, eIF4E3 acts as a tissue-specific tumor suppressor.Citation6 For instance, eIF4E3 inhibits expression of both mRNA export and translation targets of eIF4E1, consistent with its nuclear and cytoplasmic localization (). Importantly, eIF4E3 overexpression represses oncogenic transformation. Further, eIF4E3 expression is severely reduced in acute myeloid leukemia (AML) specimens and lost in HNSCC (head and neck squamous cell carcinoma), where in both cases eIF4E1 levels are known to be highly elevated. Given the potential role of eIF4E3 in suppressing eIF4E1 function, it seems likely that these cancers are driven by both the gain of oncogenic activity of eIF4E1 and loss of the suppressive activity of eIF4E3.
Figure 1. Schemes depicting the regulation of eIF4E1 activity (A) and the sequence features of eIF4E1 and eIF4E3 (B). (A) Model showing eIF4E3 competition of both nuclear and cytoplasmic functions of eIF4E1. For simplicity, only two regulatory proteins are shown (PML and 4E-BPs). The protein X on eIF4E3 represents a possible co-factor(s), which could increase its affinity for the cap in cells. The red circle denotes the cap on the 5′ end of the mRNA. In (B), asterisks highlight the regions involved in cap recognition, including the important Trp residues (W) in eIF4E1 and their corresponding residues in eIF4E3 (see main text). Black arrows on eIF4E3 show the N- and C-terminal ends for the second variant of eIF4E3. Secondary structure elements are shown by red cylinders and blue arrows for α-helices and β-strands, respectively. Note that there is ~25% identity between eIF4E1 and eIF4E3Citation5 and 100% identity between the second variant of eIF4E3 and the corresponding region of eIF4E3 variant 1.
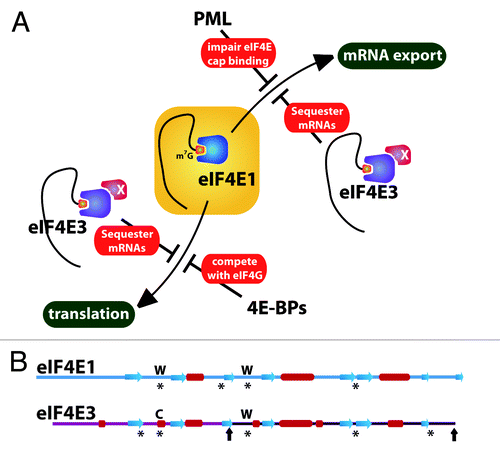
Making eIF4E3 unique among the family members is its novel recognition of the m7G cap moiety on the RNAs. For all the family members, cap-binding activity is central to their biological activities. Highlighting the importance of this activity, a cap competitor ribavirin inhibits eIF4E1 activity in AML patients leading to clinical responses.Citation7 Previous studies suggested that the only manner by which eIF4E family members recognize the m7G cap was through intercalation of the m7G cap between two aromatic residues, forming a sort of aromatic sandwich. However, initial studies of the eIF4E3 sequence revealed that it was missing one of the conserved aromatic residues to make such a sandwich.Citation5 This led to the hypothesis that eIF4E3 did not bind the m7G cap. However, our recent NMR and biophysical studies indicate that eIF4E3 does bind the cap using a unique mode of m7G cap recognition and discriminates between m7G and guanosine.Citation6 This involves an aromatic residue on one side of the cap moiety and a cluster of hydrophobic and charged residues making extensive contacts on the other side, including an important conserved cysteine residue (). This binding modality leads to weaker binding than eIF4E1, but similar to eIF4E2.
Aside from its novel cap recognition strategy, eIF4E3 displays other differences relative to eIF4E1. One example is that eIF4E3 does not associate with critical regulators of eIF4E1 function. For instance, eIF4E must bind eIF4G in order to associate with the ribosome, and, additionally, 4E-BP1 is a key negative regulator of eIF4E1.Citation4 In cells, no association is detected between either 4E-BP1 or eIF4G and eIF4E3. Biophysical studies demonstrate that binding of these factors for eIF4E3 is very much reduced relative to eIF4E1. Thus, eIF4E3 is not expected to actively engage the ribosome or compete for at least one class of inhibitor (4E-BP1). Structural analyses of the dorsal surface of eIF4E3, which is the expected binding site for these regulators, indicate that structural differences underlie the observed behavior. Another difference between eIF4E1 and eIF4E3 is that the central residue for phosphorylation-mediated regulation of eIF4E1 (S209)Citation1 is absent in eIF4E3.
Our current model is that eIF4E3 competes for the same pool of transcripts as eIF4E1, thereby impeding the ability of eIF4E1 to promote the expression of these proliferative and survival factors (). Our studies suggest that eIF4E3 impacts both mRNA export and translation.Citation6 Given the reduced affinity of eIF4E3 for the cap structure, it seems likely that additional co-factors associate with eIF4E3 in cells to increase its affinity for the cap. Alternatively (but not mutually exclusively), there could be additional elements in the RNAs themselves, such as the 4E-sensitive element (4E-SE) identified for eIF4E1,Citation3 that promote recruitment of eIF4E3. Determination of the RNA pools affected is obviously very important. Another interesting twist is the presence of a variant form of eIF4E3, which is missing the first 89 residues and, thus, many of the residues required for cap binding. It will be exciting to determine what roles this variant may play in the cell. Importantly, the tissue distribution of eIF4E3 is more limited than eIF4E1, indicating that this level of regulation is context-specific.Citation6
Many inhibitory mechanisms have been described for eIF4E1, mainly through the interaction of negative regulatory proteins such as 4E-BP1, PML and VPg with its dorsal surface.Citation4,Citation8 The mechanism proposed here is completely novel in that eIF4E3 is competing for the same pool of RNAs through a novel cap binding activity. Given that eIF4E3 impedes oncogenic transformation and its expression is lost in some cancers, this new mechanism likely has important biological and perhaps clinical implications.
References
- Graff JR, Zimmer SG. Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin Exp Metastasis 2003; 20:265 - 73; http://dx.doi.org/10.1023/A:1022943419011; PMID: 12741684
- Topisirovic I, Guzman ML, McConnell MJ, Licht JD, Culjkovic B, Neering SJ, et al. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol 2003; 23:8992 - 9002; http://dx.doi.org/10.1128/MCB.23.24.8992-9002.2003; PMID: 14645512
- Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle 2007; 6:65 - 9; http://dx.doi.org/10.4161/cc.6.1.3688; PMID: 17245113
- von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 2004; 11:503 - 11; http://dx.doi.org/10.1038/nsmb779; PMID: 15164008
- Joshi B, Lee K, Maeder DL, Jagus R. Phylogenetic analysis of eIF4E-family members. BMC Evol Biol 2005; 5:48; http://dx.doi.org/10.1186/1471-2148-5-48; PMID: 16191198
- Osborne MJ, Volpon L, Kornblatt JA, Culjkovic-Kraljacic B, Baguet A, Borden KL. eIF4E3 acts as a tumor suppressor by utilizing an atypical mode of methyl-7-guanosine cap recognition. Proc Natl Acad Sci USA 2013; 110:3877 - 82; http://dx.doi.org/10.1073/pnas.1216862110; PMID: 23431134
- Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood 2009; 114:257 - 60; http://dx.doi.org/10.1182/blood-2009-02-205153; PMID: 19433856
- Michon T, Estevez Y, Walter J, German-Retana S, Le Gall O. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J 2006; 273:1312 - 22; http://dx.doi.org/10.1111/j.1742-4658.2006.05156.x; PMID: 16519694