All living organisms must integrate nutrient and energy availability with environmental signals to coordinate their growth and development. This is at least partially achieved by the multifaceted roles of a highly conserved ancient protein kinase, target-of-rapamycin (TOR), which has been identified as a central growth regulator that modulates nutrient status and energy signaling to promote cell proliferation and growth.Citation1,Citation2 In mammals, TOR signaling has mainly been linked to amino acid sensing and insulin/growth regulator signaling. However, the mechanisms underlying responses of TOR to glucose as a universal fuel remained enigmatic in plants and animals.Citation1-Citation3
Limited evidence supports mammalian TOR signaling in direct transcriptional control. Most research has focused on the roles of TOR in translational control through phosphorylation of S6K1, as a versatile activator of translation, or eukaryotic translation initiation factor4E (eIF4E)-binding proteins (4E-BPs) to facilitate the translation of mRNAs encoding proteins promoting cell cycle progession.Citation1,Citation4 Very recently, S6K1 was reported to stimulate the synthesis of nucleotides during S-phase progress of the cell cycle through posttranslational regulation of CAD (carbamoyl-phosphate synthetase2, aspartate transcarbamoylase, dihydroorotatase), the enzyme catalyzing the first three steps of de novo pyrimidine synthesis.Citation5
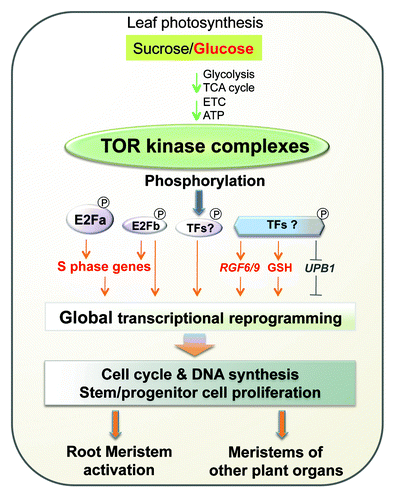
Postembryonic growth and development in plants strictly relies on glucose availability.Citation3 By exploring the regulatory mechanisms at the transition checkpoint of heterotrophic to photoautotrophic conversion in Arabidopsis seedlings, we discovered that TOR senses and transduces shoot photosynthesis-derived glucose signals through glycolysis and mitochondria energy relay to specifically control the proliferation of stem/progenitor cells in the root meristem.Citation2 Surprisingly, genome-wide expression profiling revealed that glucose-TOR signaling orchestrates global transcriptional reprogramming, which simultaneously regulates transcription, bioenergetics, metabolism, biosynthesis, signaling, folding, transport and cell cycle.Citation2 Remarkably, TOR, but not S6K1, directly phosphorylates the N terminus of E2Fa and promotes its activity in transcriptional activation of S-phase genes.Citation2 This finding uncovered a previously unrecognized TOR function in direct transcriptional regulation of cell cycle, which is beyond its well-known functions in stimulating the translation of proteins involved in cell cycle progression.Citation1
The specific phosphorylation of E2Fa by TOR kinase provided a compelling example for how glucose-TOR signaling controls transcription of S-phase genes to promote root meristem activation. However, other related E2Fs might provide partially overlapping functions in the root meristem or in the meristem of other plant organs. There are six members in the Arabidopsis E2F gene family, but only E2Fa and E2Fb share overlapping functions in binding to and activating the promoters of S-phase genes. It is likely that E2Fb is also phosphorylated and regulated by glucsoe-TOR signaling. E2Fc exhibits different expression patterns and does not appear to regulate the same cell cycle genes as E2Fa, and the atypical E2F members (E2Fd-f/DEL) were shown to exert distinct functions as repressors of gene transcription.Citation6,Citation7 Future studies will be required to determine the precise target genes and functions of different plant E2Fs. Besides E2Fs, we also revealed that glucose-TOR signaling modulated other positive and negative regulators of the root meristem activity, e.g., the upregulation of root growth factor genes (RGF6/9) and genes in the glutathione synthesis pathway required for cell cycle progression, and the downregulation of UPB1 encoding a bHLH transcription factor promoting root differentiation. The findings suggest that TOR controls a host of effectors and transcription factors for the concerted activation of cell proliferation in meristems (). Although the RGF6/9 transcripts are relatively low in the quiescent root meristems, these genes are highly activated by glucose-TOR signaling.Citation2 It will be important to investigate how TOR kinase phosphorylates and regulates the potentially distinct functions of diverse substrates in glucose-TOR signaling.
Figure 1. Model of glucose-TOR signaling in transcriptional regulation of cell cycle. Leaf photosynthesis-derived glucose is indispensable for activating TOR kinase via glycolysis and mitochondrial bioenergetic relay to control a host of effectors and transcription factors for the concerted activation of the proliferation of progenitor and stem cells in meristems. ETC, electron transport chain; RGF, root growth factors; UPB1, UPBEAT1 transcription factor; GSH, the glutathione synthesis pathway; TFs, transcription factors.
The rapid and global transcriptional reprogramming triggered by glucose-TOR signaling was not observed in animals or plants before our studies. Over the past decade, mostly long-term (18–24 h) or limited gene expression changes have been observed in mammalian cultured cells after treatments with TOR inhibitorsCitation1 or after partial reduction of TOR expression in growing plants supported by active photosynthesis.Citation8 The manifestation of gene expression changes might take longer, as the mammalian experiments were conducted at the condition with constitutively high TOR activity stimulated by insulin, serum, amino acids or tsc (tuberous-sclerosis-complex) mutants when TOR inhibitors were applied. The unique experimental design using the 3-d-after-germination seedlings at the heterotrophic to photoautotrophic transition checkpoint, exhibiting normal growth and development in the absence of photosynthesis or exogenous sugars, offered a low TOR signaling background to observe rapid, dynamic and clear glucose induction in global transcriptional changes.Citation2 Therefore, it appears crucial to probe the rapid changes in TOR-mediated transcriptome in a bioenergetically quiescent state with minimal TOR signaling background before adding specific TOR-stimulating signals and TOR inhibitors. Cell cycle regulation occurs mostly in meristems of intact plants, enriching the meristem tissues and cells for expression profiling is also critical to reveal the clear involvement of glucose-TOR signaling in the control of S-phase genes. As glucose is a universal fuel and metabolic/biomass precursor for most cells, the unexpected findings in plants may illuminate future research on novel and fundamental regulatory mechanisms in glucose-TOR signaling in plants, animals and humans, which share complexity in cell cycle controls by nutrient and energy signaling through communication and coordination among different organs.
References
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274 - 93; http://dx.doi.org/10.1016/j.cell.2012.03.017; PMID: 22500797
- Xiong Y, McCormack M, Li L, Hall Q, Xiang CB, Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013; 496:181 - 6; http://dx.doi.org/10.1038/nature12030; PMID: 23542588
- Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem 2012; 287:2836 - 42; http://dx.doi.org/10.1074/jbc.M111.300749; PMID: 22134914
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010; 328:1172 - 6; http://dx.doi.org/10.1126/science.1187532; PMID: 20508131
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 2013; 339:1323 - 8; http://dx.doi.org/10.1126/science.1228792; PMID: 23429703
- de Jager SM, Scofield S, Huntley RP, Robinson AS, den Boer BG, Murray JA. Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol 2009; 71:345 - 65; http://dx.doi.org/10.1007/s11103-009-9527-5; PMID: 19662336
- Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol 2009; 19:111 - 8; http://dx.doi.org/10.1016/j.tcb.2009.01.002; PMID: 19201609
- Dobrenel T, Marchive C, Azzopardi M, Clement G, Moreau M, Sormani R, et al. Sugar metabolism and the plant target of rapamycin kinase:asweetoperaTOR. Frontiers in Plant Biology 2013; doi: http://dx.doi.org/10.3389/fpls.2013.00093