Abstract
Tumor suppressor p53 maintains genome stability by differentially activating target genes that control diverse cellular responses, such as the antioxidant response, cell cycle arrest and apoptosis. Despite the fact that many p53 downstream genes have been well characterized, novel p53 target genes are continuously being identified. Here, we report that Tpt1 is a direct target gene of p53. We found that p53 upregulates the transcription of Tpt1 and identified a p53-responsive element in the promoter of the mouse Tpt1 gene. Furthermore, p53-dependent induction of Tpt1 was able to reduce oxidative stress, minimize apoptosis, and promote cell survival in response to H2O2 challenge. In addition, a positive correlation between the expression of p53 and Tpt1 only existed in normal lung tissues, not in lung tumors. Such positive correlation was also found in lung cell lines that contain wild-type p53, but not mutated p53. Based on the important role of Tpt1 in cancer development, chemoresistance, and cancer reversion, identification of Tpt1 as a direct target gene of p53 not only adds to the complexity of the p53 network, but may also open up a new avenue for cancer prevention and intervention.
Introduction
Tumor protein translationally controlled 1 (Tpt1), also known as translationally controlled tumor protein (TCTP), is an evolutionary conserved protein that shares a high degree of homology from plants to mammals, implicating its fundamental functions.Citation1,Citation2 It is ubiquitously expressed in many tissues and cell types, and homozygous knockdown of Tpt1 is embryonic lethal.Citation3 Tpt1 is composed of three domains, known as the: 1) β-stranded core domain, 2) flexible loop domain, and 3) helical domain. The flexible loop and the helical domains are specific for Tpt1. Currently, the helical domain, which contains a tubulin-binding regionCitation4 and a Ca2+-binding area,Citation5 is the only region that has been shown to affect the molecular function of Tpt1.
Currently, the cellular function and molecular regulation of Tpt1 are uncertain, even though its possible participation in diverse biological processes has been reported. As a cellular stress biomarker, the expression of Tpt1 was upregulated in response to mild H2O2, thermal shock, or genotoxic reagents, which confers protection against cell death.Citation8,Citation9 For instance, Tpt1 was shown to act as a heat-shock protein that binds denatured proteins and protects them from thermal shock.Citation8 Tpt1 is inversely associated with the sensitivity of cells to H2O2.Citation9 In addition, Tpt1 is thought to promote cell survival by enhancing the anti-apoptotic response and suppressing proapoptotic activities.Citation9-Citation11
Tpt1 is associated with cancer progression and is highly expressed in many tumors.Citation10 In a cultured colon cancer cell line, knockdown of Tpt1 was shown to inhibit cell proliferation, migration, and invasion.Citation12 In vivo, knockdown of Tpt1 reduced tumor growth and liver metastasis when the colon cancer cells were injected into nude mice.Citation12 Similarly, siRNA-mediated downregulation of Tpt1 in prostate cancer cells inhibited cell growth and enhanced apoptosis through activation of caspase-8 and caspase-3.Citation10 Another interesting observation is that Tpt1 is downregulated at both mRNA and protein levels during tumor reversion.Citation13-Citation15 The anti-apoptotic activity of Tpt1 was also demonstrated by the fact that ectopic overexpression of Tpt1 protects HeLa and U2OS cells from cell death in response to etoposide treatment (Tpt1 was referred as fortilin in this report).Citation16 Collectively, the existing experimental results highlight the important role of Tpt1 not only in tumor development and regression, but also in dictating chemoresistance. Therefore, Tpt1 may represent a drug target for cancer prevention and intervention.
Tumor suppressor p53 exerts its function by transactivating an array of target genes involved in several important cellular processes, such as cell redox maintenance, cell cycle checkpoints, cellular senescence, proliferation, and apoptosis.Citation6,Citation7 Previously, we reported that the antioxidant function of p53 results from crosstalk of p21 with the Nrf2-Keap1-ARE pathway, which regulates the major cellular antioxidant response.Citation17,Citation18 When cells are under genotoxic stress, p53 upregulates targets genes that are involved in cell cycle arrest and DNA repair to maintain the integrity of the genome. In extreme cases, in which damage is beyond repair, p53 directs cells to commit suicide by activating the apoptotic response. Therefore, p53 is a guardian of the genome and p53 is crucial to maintain the normal function of cells. The importance of p53 in cancer is accentuated by the fact that more than 50% of human tumors contain a mutated and non-functional p53. Functioning as a sensor of various stresses, p53 can be activated by nongenotoxic stress (hypoxia, heat shock, redox changes, nutrient depression), genotoxic stress (including UV, radiation, carcinogens, and cytotoxic drugs), and oncogenic stress. It is believed that different types of stress activate a distinct set of p53-target genes to cope with the particular stress.
Many p53-target genes, including p21, Bax, PUMA, Bcl2, ERCC2/3, and 14-3-3s, have been well characterized. Nevertheless, new target genes are continuously being identified and their functions delineated. Tumor protein 53-induced nuclear protein 1 (TP53INP1), the G protein-coupled receptor 87 (GPR87), syntaxin 6, and BNIP3 are among these newly identified p53 target genes that have various biological functions. p53-mediated upregulation of TP53INP1 enhances cellular antioxidant capacity.Citation19 GPR87 promotes cell survival under genotoxic stress, and knockdown of GPR87 sensitizes cancer cells to DNA damage reagents.Citation20 Syntaxin 6, a vesicular transporter protein, modulates cell adhesion and enhances cell survival.Citation21 In another report, p53 was shown to be upregulated to protect cells from hypoxia-induced cell death by negatively regulating its target gene BNIP3, a member of the BCL2 family with pro-apoptotic function.Citation22
As indicated by its name, Tpt1 was previously recognized as a tumor-associated protein that is primarily regulated at the level of protein translation. Recently, Tpt1 was found to interact with p53 and to negatively regulate p53 by increasing its degradation.Citation23 Consistent with these findings, another study determined that, indeed, Tpt1 promotes p53 degradation by inhibiting MDM2-mediated ubiquitination and degradation of p53.Citation24 However, the precise function and the mechanistic regulation of Tpt1 remain elusive. In this report, we identified Tpt1 as a novel p53 target gene, and a unique p53-responsive element was identified within the 1 kb promoter region of mouse Tpt1. Combined with the published data, our results indicate the existence of a negative feedback mechanism controlling the expression of both p53 and Tpt1. Identification of Tpt1 as a direct target gene of p53 not only adds to the complexity of the p53 network, but may also open up a new avenue for cancer prevention and intervention.
Results
Identification of a p53-response element in the promoter of Tpt1
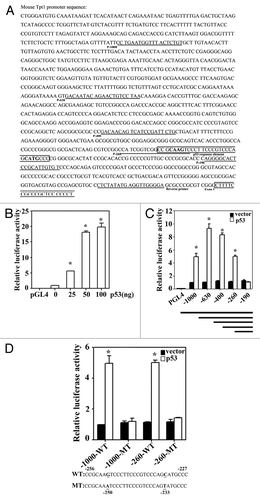
The 1 kb sequence of the mouse Tpt1 promoter upstream of the beginning of exon 1 is shown in . Examination of this promoter sequence yielded several putative p53-response elements. Next, the 1 kb promoter was inserted into the cloning site of a luciferase reporter vector and a luciferase reporter gene assay was performed in HCT116-p53−/− cells transfected with different amounts of p53. The luciferase activity significantly increased in a dose-dependent manner with a maximum induction of 20-fold (). To further define p53-response elements, a total of 5 fragments containing different sizes of the mouse Tpt1 promoter were individually cloned into a luciferase reporter vector (, lines), and the promoter activity of each fragment was measured in HCT116-p53−/− cells co-transfected with an empty vector or a p53 expression vector (). p53 significantly enhanced luciferase activities in all the reporters containing 260 bp or more of the Tpt1 promoter, but not the one with 190 bp (). Close examination of the promoter sequence between 190–260 bp led to the identification of a putative p53-response element from −227 to −256 of the mouse Tpt1 promoter (The boxed sequence in ). This putative p53-response element (WT) and its mutated sequence (MT) were cloned in the context of either the 1 kb or 260 bp of the promoter, and reporter gene assay was performed (). Indeed, luciferase activities under the control of the WT sequence were markedly induced by overexpression of p53 (). However, mutation of 2 nucleotides (C to T at −233; G to A at −250) completely abolished the upregulation of luciferase activity by p53 (). This result indicates that the sequence between −227 bp to −256 bp of the mouse Tpt1 promoter contains an authentic p53-response element.
Figure 1. Identification of a p53-response element in the promoter of Tpt1.(A) The 1 kb DNA sequence of the mouse Tpt1 promoter. The DNA primers used to amplify 1 kb, 630 bp, 400 bp, 260 bp, and 190 bp of the mouse Tpt1 promoter fragments are underlined and labeled as P-1000, P-630, P-400, P-260, and P-190 (forward primers). The common reverse primer is also underlined and labeled. The putative p53-response element and the beginning of exon 1 are boxed and labeled. (B) Ectopic expression of p53 in HCT116-p53−/− cells increased Tpt-1 promoter-dependent firefly luciferase activities. The mouse Tpt1 promoter (1 kb) was cloned upstream of the luciferase reporter gene vector. The construct was co-transfected into HCT116-p53−/− cells growing in 24-well plates, with the indicated amounts of a p53-expression plasmid. A control vector pGL4, which only contains a minimal promoter, was included as a negative control. (C) A p53-response element in the mouse Tpt1 promoter was mapped to the sequence within −190 to −260 nt. The different sizes of the mouse Tpt1 promoter were cloned upstream of the luciferase reporter gene vector (see line draws at the bottom of the panel, also see for the primer pair information). These constructs were co-transfected into HCT116-p53−/− cells alone with a control vector or a p53 expression vector. Luciferase activities were measured. (D) Identification of the p53-response element. The p53-response element in the promoter of mouse Tpt1 (WT) is shown at the bottom of the panel. Two highly conserved bases (underlined) were mutated in MT. The fragment containing the putative p53-response element (WT) or the mutated one (MT) in the context of either 1 kb or 260 bp was cloned into the luciferase reporter vector. The luciferase activities of these reporters in the absence or presence of transfected p53 were measured.
p53 regulates basal and induced levels of Tpt1
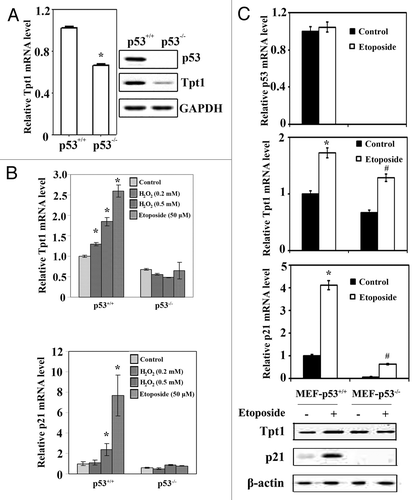
Next, the basal expression of Tpt1 at both mRNA and protein levels was compared in HCT116-p53+/+ and HCT116-p53−/− cells (). Knockout of p53 in HCT116-p53−/− cells was confirmed by immunoblot analysis with an anti-p53 antibody (, right panel). HCT116-p53−/− cells had a reduced mRNA and protein level of Tpt1 compared with HCT116-p53+/+ cells (). This result indicates that p53 controls the basal expression of Tpt1. To further demonstrate the important role of p53 in transactivating Tpt1 in response to genotoxic stress, HCT116-p53+/+ and HCT116-p53−/− cells were treated with H2O2 and etoposide. H2O2 and etoposide induced Tpt1 mRNA expression only in HCT116-p53+/+, not in HCT116-p53−/− cells (, upper panel). p21, the classical target gene of p53, was included as a positive control (, lower panel). The induction pattern of Tpt1 transcript is similar to p21, although to a lesser extent in response to DNA damaging reagents. This result further supports the finding that Tpt1 is a p53 target gene. In order to eliminate any caveats that may be associated with using cancer cell lines, primary MEF cells derived from a p53+/+ or p53−/− mouse embryo were compared for the expression of Tpt1 under both basal and induced conditions. The status of MEF-p53 was confirmed by the mRNA level of p53 (). The mRNA level of Tpt1 or p21 was induced by etoposide in both MEF-p53+/+ or MEF-p53−/− cells, although it was lower in MEF-p53−/− cells than MEF-p53+/+ cells (). Interestingly, the basal protein level of Tpt1 was not lower in MEF-p53−/− than that in MEF-p53+/+ cells, whereas there were no detectable levels of p21 in MEF-p53−/− cells (). In response to etoposide, the protein level of Tpt1 was only induced in MEF-p53+/+ cells, not in MEF-p53−/− cells (). Taken together, these results clearly demonstrate that p53 controls the expression of Tpt1. Nevertheless, p53-mediated regulation of Tpt1 expression seems to be weaker than that of p21. This may implicate that the p53-response element in the promoter of Tpt1 is controlled less tightly by p53 compared with that in the promoter of p21.
Figure 2. p53 upregulates basal and induced levels of Tpt1.(A) The mRNA and protein levels of Tpt1 were elevated in HCT116-p53+/+ cells, compared with HCT116-p53−/− cells. Tpt1 expression at both the mRNA level and the protein level in HCT116-p53+/+ and HCT116-p53−/− cells was compared using quantitative real-time RT-PCR (left panel) and immunoblot analysis (right panel). (B) Transcription of Tpt1 is enhanced by H2O2 or etoposide in HCT116-p53+/+, not HCT116-p53−/− cells. The mRNA level of Tpt1 (middle panel) and p21 (bottom panel) was examined by qRT-PCR in HCT116-p53+/+ and HCT116-p53−/− cells treated with two doses of H2O2 or etoposide for 24 h. (C) Tpt1 expression is controlled by p53 in the primary MEF cells. MEF-p53+/+, and MEF-p53−/− cells were left untreated or treated with 25 μM etoposide for 16 h. The basal and induced expression of endogenous Tpt1 and p21 at both mRNA and protein levels were compared.
Tpt1 maintains cellular redox balance and minimizes cell death in response to H2O2
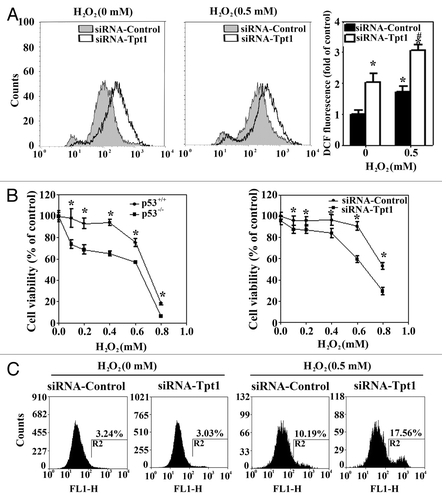
The role of Tpt1 in protecting against cellular toxicity was explored in HCT116-p53+/+ cells transfected with a control siRNA or a siRNA specific for Tpt1. Knockdown of Tpt1 was sufficient to tip the redox balance toward an increase in oxidative stress under basal conditions (). Treatment with H2O2 further enhanced the level of ROS with a higher level of ROS observed in cells transfected with siRNA-Tpt1 (). Next, cellular toxicity was measured by two independent methods, MTT and apoptosis assays. Similar to the cell viability curves between HCT116-p53−/− and HCT116-p53−/− cells, cells transfected with siRNA-Tpt1 were more sensitive to H2O2 compared with siRNA-control transfected cells (). In an annexin V-based apoptosis assay, knockdown of Tpt1 had no effect on cell viability on its own but significantly enhanced H2O2-mediated apoptotic cell death (). Collectively, these results indicate that Tpt1 confers a cellular-protective response.
Figure 3. Tpt1 is required for maintaining cellular redox balance and cell survival in response to H2O2. (A) Suppression of Tpt1 expression increases the level of ROS. HCT116-p53+/+ cells were transfected with siRNA-Tpt1 or siRNA-control. At 48 h post-transfection cells were either left untreated or treated with H2O2 for 24 h, intracellular ROS levels were measured by DCF fluorescence dye/flow cytometry. (B) Suppression of Tpt1 expression decreases cell viability in response to H2O2. HCT116-p53+/+, HCT116-p53−/−, HCT116-p53+/+ transfected with siRNA-control or siRNA-Tpt1 were treated with the indicated doses of H2O2 for 48 h. Cell viability was measured by MTT assay at 72 h post-transfection. (C) Suppression of Tpt1 expression enhances apoptotic cell death. HCT116-p53+/+ cells transfected with siRNA-control or siRNA-Tpt1 were either left untreated or treated with 0.5 mM H2O2 for 24 h. Apoptotic cells were measured using Annexin V-FITC/flow cytometry at 72 h post-transfection.
Association between p53 and Tpt1 expression in normal vs. tumor lung tissues and lung cell lines
Next, the correlation between the expression of p53 and Tpt1 in lung tissues or cell lines was analyzed. A tissue microarray containing both normal lung (89 patients) and cancerous tissues (80 patients) was subjected to immunohistochemistry (IHC) staining with anti-p53 and anti-Tpt1 antibodies. The correlation in their expression was statistically calculated using an expression index. In normal tissues, a positive correlation between the expression of p53 and Tpt1 was observed (r = 0.707, P < 0.01), although p53 expression was relatively weak in normal tissues compared with tumor tissues (). Inversely, no correlation was found in their gene expression in tumor tissues (r = 0.207, P > 0.05) (). Since it has been well documented that more than 50% of lung cancer tissues contain a mutated or non-functional p53, the IHC result implicates that a positive correlation in the expression between p53 and Tpt1 is only found in tissues that contain a functional p53.
Figure 4. There is a positive correlation in the expression of a functional p53 and Tpt1. (A) Positive correlation between p53 and Tpt1 only exists in normal but not tumor tissues. Lung tissue microarrays were subjected to H&E staining and IHC analysis with anti-p53 and anti-Tpt1 antibodies. Six representative slices from normal or tumor tissues are presented. In normal tissues, p53 and Tpt1 showed a similar staining pattern, indicating a positive correlation in their expression (, panels 1–6). Different types of lung cancer tissues were presented in panel 7–12. Positive correlation: 7, adenocarcinoma; 8, squamous cell carcinoma. Negative correlation: 9, squamous cell carcinoma; 10, small cell lung cancer; 11, squamous cell carcinoma; and 12, adenocarcinoma. (B) The level of wild-type p53 correlates with the level of Tpt1. Protein levels of p53, Tpt1, and p21 was analyzed in 8 lung cell lines by immunoblot analysis. The status of p53 of each cell line is labeled. HBE, H292, A549, and H838 cells contain wild-type p53; H69 and Calu-1 are p53-null; and H1703 and H23 have p53 mutated.
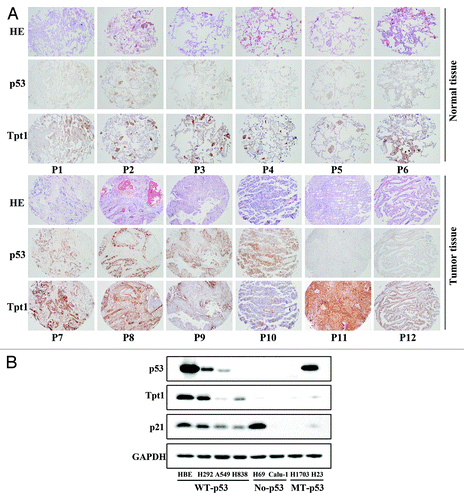
Next, we examined the protein levels of p53 and Tpt1 in 8 established lung cell lines where the status of p53 has previously been defined: HBE, H292, A549, and H838 cells contain wild-type p53; H69 and Calu-1 are p53-null; and H1703 and H23 have p53 mutated. p21 was included as a positive control. In cell lines bearing a wild-type p53 (HBE, H292, A549, and H838), a general positive correlation between p53 and Tpt1, as well as p53 and p21 were observed (). A slight deviation from the positive correlation between p53 and Tpt1 was observed in H838 cells, where p53 was under the detection limit but Tpt1 showed a band that is more intense than that in A549, H69, and Calu-1 (). This likely reflects the difference in the expression of other transcription factors that control the promoter activity of Tpt1, since these cell lines were derived from different individuals. This notion is further supported by the huge discrepancy among the level of p53 and p21 in H69, where the expression of p21 is high even though p53 is not expressed (). In H69 and Calu-1 cells that do not contain p53, only a low basal level of Tpt1 was detected (). Conversely, in p53-mutated H1703 and H23 cell lines, Tpt1 and p21 expressed at very low levels in both cell lines, although expression of p53 is much higher in H23 compared with that in H1703. These results demonstrate a potential positive correlation between wild-type p53 and Tpt-1.
Discussion
The tumor suppressor p53 is a well-known transcription factor that modulates numerous genes involved in many cellular functions such as cell cycle arrest, induction of apoptosis, and anti-angiogenesis. The biological functions of p53 depend on many factors, and are mediated largely through the actions of its downstream target genes. Although several p53 target genes have been characterized, many others remain to be identified. The identification of p53 target genes is important for understanding the mechanism by which p53 mediates its biological role in normal and cancerous cells.
In our current study, we demonstrated that p53 upregulates Tpt1, a protein that has been shown to be involved in cancer progression and chemoresistance. Previously, other p53 target genes have been identified, including p21, Bax, PUMA, Bcl 2, ERCC2/3, and 14-3-3s. These genes have various biological functions. p53-dependent activation of Tpt1 decreased oxidative stress and apoptosis in several different types of cancer cells, whereas knockdown of Tpt1 using siRNA enhanced the levels of reactive oxygen species and sensitized cells to H2O2-induced cell death. Interestingly, during the course of this study Amson et al., determined that p53 binds to a p53-responsive element in the Tpt1 promoter (−384 to −200), leading to the transcriptional repression of Tpt1 (Fig. 1 of the paper published in “Nature Medicine”Citation24). We therefore cloned several fragments (up to 1500 bp) of the human Tpt1 promoter into the luciferase reporter vector. However, none of the human Tpt1 promoter reporters showed a reduced luciferase activity by overexpression of p53 in HCT116-p53−/− cells, where mouse Tpt1 promoter reporter (1 kb) was induced dramatically under the same condition (data not shown). The discrepancy between their study and ours is unclear. It seems that the p53-mediated repression of Tpt1 reported in the literature is exclusively from the same group. On the other hand, in another study, stable knockdown of p53 in a nasopharyngeal carcinoma cell line overexpressing wild-type p53 resulted in a more than 2-fold decrease in Tpt1 expression at the protein level,Citation25 supporting our finding that Tpt1 is a target gene of p53.
The identification of Tpt1 as a direct target gene of p53 increases our understanding of the physiological roles of wild-type p53, as well as the pathological effects of p53 mutations in human carcinogenesis. Since Tpt1 plays an important role in cancer development, chemoresistance, and cancer reversion, our findings may also open up a new avenue for cancer prevention and intervention.
Materials and Methods
Cell culture, p53+/− mice, and primary MEF cell isolation
H292, A549, H838, H69, Calu-1, H1703, H23, HBE, HCT116-p53+/+, and HCT116-p53−/− cells were maintained in DMEM medium with 10% FBS. p53+/− mice were purchased from the Jackson Laboratory. p53+/+ and p53−/− mice were obtained by breeding p53+/− mice and confirmed by genotyping. Primary MEF cells were isolated from 13 p.c. embryos and cultured in DMEM. MEF-p53+/+ and MEF-p53−/− cells used in this study are passage number 3.
Plasmids
Flag-p53 in pCDNA3 was purchased from Addgene. The 1 kb fragment of the Tpt1 promoter and several deletion fragments were amplified by PCR using a mouse genomic BAC library (BACPAC, Children’s Hospital Oakland research Institute) as the template, and then cloned into the pGL4 vector between the Xho I and Hind III sites. PCR primers include 5 forward primers, and 1 common reverse primer is shown in .
Luciferase reporter gene assay
A Tpt1 promoter-luciferase construct and a rellina luciferase expression vector were co-transfected into HCT116-p53−/− cells with p53 plasmid or empty vector. The luciferase activity was measured 48 h after transfection using the dual-luciferase assay (Promega).
Transfection
Transfection of cDNA was performed with Lipofectamine Plus (Invitrogen). The validated siRNA-Tpt1 and siRNA-control were purchased from Qiagen. HiPerfect transfection reagent (Qiagen) was used to deliver siRNA.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using Trizol (Invitrogen). cDNA was generated using reverse transcriptase (Promega). qRT-PCR was performed using Sybr Green PCR master mix (ABI).
The primers for p21 expression analysis are:
5′-GACACCACTGGAGGGTGACT(F)
5′-CAGGTCCACATGGTCTTCCT (R)
The primers for Tpt1 expression analysis are:
5′-AAATGTTAACAAATGTGGCAATTAT(F)
5′-AACAATGCCTCCACTCCAAA(R).
The primers for GAPDH are:
5′-GCTCATTTCCTGGTATGACAACG(F)
5′-AGGGGTCTACATGGCAACTG (R).
GAPDH mRNA level was used as an internal control. The mRNA level of Tpt1 or p21 was normalized to its GAPDH and the value in the untreated sample of HCT116-p53+/+ was set to 1.
ROS detection, cell viability (MTT), and cell-death assays
To detect ROS, cells were treated with H2O2 for 24 h and incubated with dichlorofluorescein (DCF) (Sigma) before measuring ROS by flow cytometry. Cell viability was measured by 3-(4, 5 dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) method (Sigma). Apoptotic cells were detected using the Annexin V-FITC apoptosis detection kit (Sigma) in combination with flow cytometry. All procedures have been reported in detail previously.Citation17
Tissue microarray, IHC staining, and expression index
Paraffin embedded tissue microarray slides, purchased from Xi’an Alena Biotechnology Co, Ltd were analized by hematoxylin-eosin (HE) staining, and IHC staining for p53 and Tpt1 was performed according to our publiched procedures.Citation26 All slides were evaluated in a blinded manner by two pathologists for the intersity of staining (I value from 0–3) and the percentage of stained area (P value from 0–90%). Expression index was calculated according to the following formula: Index = I + 10 × P. Four catagories: no expression (index score = 0), weak expression (index score = 1–4), moderate expression (index score-5–8), and strong expression (index score = 9–12), were used for Spearman correlation analysis.
Statistical test
The data were analyzed using the SPSS software (version 13.0; SPSS Inc). Spearman correlation was used to analyze the relationship between the expression of p53 and Tpt1 in normal vs. tumor lung tissues. All P values were two-tailed. The statistical significance was defined as P < 0.05. For luciferase reporter assay, real-time PCR, and MTT assays, experiments were repeated 3 times, each with duplicate or triplicate samples. Results are expressed as mean ± SD. One-way ANOVA was used to compare the means among groups.
Acknowledgments
This work was supported by the NIH grants 2R01ES015010 and R01CA154377 to DD Z, and R01AI079056 to DF.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M. Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci USA 2010; 107:16384 - 9; http://dx.doi.org/10.1073/pnas.1007926107; PMID: 20736351
- Sanchez JC, Schaller D, Ravier F, Golaz O, Jaccoud S, Belet M, et al. Translationally controlled tumor protein: a protein identified in several nontumoral cells including erythrocytes. Electrophoresis 1997; 18:150 - 5; http://dx.doi.org/10.1002/elps.1150180127; PMID: 9059837
- Chen SH, Wu PS, Chou CH, Yan YT, Liu H, Weng SY, et al. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol Biol Cell 2007; 18:2525 - 32; http://dx.doi.org/10.1091/mbc.E07-02-0188; PMID: 17475776
- Gachet Y, Tournier S, Lee M, Lazaris-Karatzas A, Poulton T, Bommer UA. The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J Cell Sci 1999; 112:1257 - 71; PMID: 10085260
- Kim M, Jung Y, Lee K, Kim C. Identification of the calcium binding sites in translationally controlled tumor protein. Arch Pharm Res 2000; 23:633 - 6; http://dx.doi.org/10.1007/BF02975253; PMID: 11156187
- Demidenko ZN, Korotchkina LG, Gudkov AV, Blagosklonny MV. Paradoxical suppression of cellular senescence by p53. Proc Natl Acad Sci USA 2010; 107:9660 - 4; http://dx.doi.org/10.1073/pnas.1002298107; PMID: 20457898
- Johansson H, Vizlin-Hodzic D, Simonsson T, Simonsson S. Translationally controlled tumor protein interacts with nucleophosmin during mitosis in ES cells. Cell Cycle 2010; 9:2160 - 9; http://dx.doi.org/10.4161/cc.9.11.11841; PMID: 20505363
- Gnanasekar M, Dakshinamoorthy G, Ramaswamy K. Translationally controlled tumor protein is a novel heat shock protein with chaperone-like activity. Biochem Biophys Res Commun 2009; 386:333 - 7; http://dx.doi.org/10.1016/j.bbrc.2009.06.028; PMID: 19523440
- Lucibello M, Gambacurta A, Zonfrillo M, Pierimarchi P, Serafino A, Rasi G, et al. TCTP is a critical survival factor that protects cancer cells from oxidative stress-induced cell-death. Exp Cell Res 2011; 317:2479 - 89; http://dx.doi.org/10.1016/j.yexcr.2011.07.012; PMID: 21801721
- Gnanasekar M, Thirugnanam S, Zheng G, Chen A, Ramaswamy K. Gene silencing of translationally controlled tumor protein (TCTP) by siRNA inhibits cell growth and induces apoptosis of human prostate cancer cells. Int J Oncol 2009; 34:1241 - 6; PMID: 19360337
- Liu H, Peng HW, Cheng YS, Yuan HS, Yang-Yen HF. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol Cell Biol 2005; 25:3117 - 26; http://dx.doi.org/10.1128/MCB.25.8.3117-3126.2005; PMID: 15798198
- Ma Q, Geng Y, Xu W, Wu Y, He F, Shu W, et al. The role of translationally controlled tumor protein in tumor growth and metastasis of colon adenocarcinoma cells. J Proteome Res 2010; 9:40 - 9; http://dx.doi.org/10.1021/pr9001367; PMID: 19621893
- Telerman A, Amson R. The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat Rev Cancer 2009; 9:206 - 16; http://dx.doi.org/10.1038/nrc2589; PMID: 19180095
- Tuynder M, Fiucci G, Prieur S, Lespagnol A, Géant A, Beaucourt S, et al. Translationally controlled tumor protein is a target of tumor reversion. Proc Natl Acad Sci USA 2004; 101:15364 - 9; http://dx.doi.org/10.1073/pnas.0406776101; PMID: 15489264
- Tuynder M, Susini L, Prieur S, Besse S, Fiucci G, Amson R, et al. Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Proc Natl Acad Sci USA 2002; 99:14976 - 81; http://dx.doi.org/10.1073/pnas.222470799; PMID: 12399545
- Li F, Zhang D, Fujise K. Characterization of fortilin, a novel antiapoptotic protein. J Biol Chem 2001; 276:47542 - 9; http://dx.doi.org/10.1074/jbc.M108954200; PMID: 11598139
- Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell 2009; 34:663 - 73; http://dx.doi.org/10.1016/j.molcel.2009.04.029; PMID: 19560419
- Rotblat B, Melino G, Knight RA. NRF2 and p53: Januses in cancer?. Oncotarget 2012; 3:1272 - 83; PMID: 23174755
- Cano CE, Gommeaux J, Pietri S, Culcasi M, Garcia S, Seux M, et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res 2009; 69:219 - 26; http://dx.doi.org/10.1158/0008-5472.CAN-08-2320; PMID: 19118006
- Zhang Y, Qian Y, Lu W, Chen X. The G protein-coupled receptor 87 is necessary for p53-dependent cell survival in response to genotoxic stress. Cancer Res 2009; 69:6049 - 56; http://dx.doi.org/10.1158/0008-5472.CAN-09-0621; PMID: 19602589
- Zhang Y, Shu L, Chen X. Syntaxin 6, a regulator of the protein trafficking machinery and a target of the p53 family, is required for cell adhesion and survival. J Biol Chem 2008; 283:30689 - 98; http://dx.doi.org/10.1074/jbc.M801711200; PMID: 18779328
- Feng X, Liu X, Zhang W, Xiao W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J 2011; 30:3397 - 415; http://dx.doi.org/10.1038/emboj.2011.248; PMID: 21792176
- Rho SB, Lee JH, Park MS, Byun HJ, Kang S, Seo SS, et al. Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett 2011; 585:29 - 35; http://dx.doi.org/10.1016/j.febslet.2010.11.014; PMID: 21081126
- Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, et al. Reciprocal repression between P53 and TCTP. Nat Med 2012; 18:91 - 9; http://dx.doi.org/10.1038/nm.2546; PMID: 22157679
- Sun Y, Yi H, Zhang PF, Li MY, Li C, Li F, et al. Identification of differential proteins in nasopharyngeal carcinoma cells with p53 silence by proteome analysis. FEBS Lett 2007; 581:131 - 9; http://dx.doi.org/10.1016/j.febslet.2006.12.008; PMID: 17184779
- Zheng H, Whitman SA, Wu W, Wondrak GT, Wong PK, Fang D, et al. Therapeutic potential of Nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes 2011; 60:3055 - 66; http://dx.doi.org/10.2337/db11-0807; PMID: 22025779