Abstract
Understanding transcriptional changes during cancer progression is of crucial importance to develop new and more efficacious diagnostic and therapeutic approaches. It is well known that ErbB2 is overexpressed in about 25% of human invasive breast cancers. We have previously demonstrated that p130Cas overexpression synergizes with ErbB2 in mammary cell transformation and promotes ErbB2-dependent invasion in three-dimensional (3D) cultures of human mammary epithelial cells. Here, by comparing coding and non-coding gene expression profiles, we define the invasive signatures associated with concomitant p130Cas overexpression and ErbB2 activation in 3D cultures of mammary epithelial cells. Specifically, we have found that genes involved in amino acids synthesis (CBS, PHGDH), cell motility, migration (ITPKA, PRDM1), and angiogenesis (HEY1) are upregulated, while genes involved in inflammatory response (SAA1, S100A7) are downregulated. In parallel, we have shown that the expression of specific miRNAs is altered. Among these, miR-200b, miR-222, miR-221, miR-R210, and miR-424 are upregulated, while miR-27a, miR-27b, and miR-23b are downregulated. Overall, this study presents, for the first time, the gene expression changes underlying the invasive behavior following p130Cas overexpression in an ErbB2 transformed mammary cell model.
Introduction
Breast cancer is the leading cause of cancer-related death in women world-wide.Citation1 Despite significant advances in breast cancer diagnosis and treatment, several major unresolved clinical and scientific problems remain, such as the understanding of the causes of tumor progression and recurrence and how to predict them.Citation2
ErbB2 is a well-known oncoprotein that belongs to the epidermal growth factor receptor (EGFR) family. It is overexpressed in approximately 25% of invasive breast cancers.Citation3 In particular, overexpression of ErbB2 has been demonstrated to promote breast cancer invasion and metastasis and to correlate with poor patient survival.Citation4-Citation7 However, ErbB2 is also overexpressed in non-invasive mammary ductal carcinomas in situ (DCIS).Citation8 Indeed, ErbB2 amplification or overexpression seems to be crucial but not sufficient for the transition from in situ to invasive cancer, and additional hits are required for the progression of ErbB2-positive tumors. Although the molecular and genetic events underlying ErbB2-positive tumor invasion and metastasis are still not well understood, intense investigation has led to the notion that molecules involved in cell adhesion and migration are critical in this process.Citation9 Moreover, it has recently been reported that altered lysosomal biogenesis and distribution play a key role in ErbB2-dependent cancer cell migration, invasion, and metastasis.Citation10
The adaptor protein p130Cas (Crk-associated substrate) has been extensively reported to act as a scaffold molecule for signaling platforms that drive cell migration and invasion by regulating intracellular cytoskeletal and effector proteins in response to extracellular matrix and growth factor signals.Citation11 We have previously demonstrated that p130Cas is a crucial regulator of ErbB2-induced mammary tumorigenesis. In particular, p130Cas is required for ErbB2 signal transduction, in vivo tumor growth and lung colonization.Citation12 By using 3D cultures of MCF10A cells modified by Muthuswamy et al.Citation13 to express a chimeric ErbB2 receptor, which can be activated by the synthetic ligand AP1510 (MCF10A.B2 cells), we demonstrated that the concomitant overexpression of p130Cas and ErbB2 activation confer invasive properties to 3D mammary acini. Indeed, in 3D culture conditions, MCF10A.B2 cells form acini-like spheroids, that recapitulate the terminal ductal lobular unit of human adult breast. Activation of ErbB2 disrupts the morphogenetic process and results in the formation of multiacinar, non-invasive structures, modeling the early stages of breast cancer onset.Citation13 In contrast, MCF10A.B2 cells that overexpress p130Cas give rise to multiacinar structures that, upon stimulation with AP1510, acquire invasive protrusions.Citation12,Citation14 This invasive phenotype is characterized by activation of Erk1/2 MAPKs and PI3K/Akt pathways that, in turn, lead to activation of distinct downstream effectors like mTOR/p70S6K and Rac1, respectively.Citation14 The clinical relevance of these findings is supported by the in silico analysis indicating that patients with breast cancers with elevated expression of both p130Cas and ErbB2 have a higher risk of developing distant site metastasis compared with patients with low levels of p130Cas.Citation14 Overall, these data highlight the importance of p130Cas and ErbB2 cooperation in promoting breast cancer progression.
Nothing is known about gene expression changes underlying the transition from non-invasive to invasive phenotype in the presence of p130Cas overexpression and ErbB2 activation. Emerging technologies, such as coding and non-coding gene microarrays, have greatly improved the capability to identify key molecular targets involved in a specific cellular process. In particular, the past decade has shown remarkable advances in our knowledge of microRNAs and their functional importance in cancer predisposition, initiation, and progression, with several small non-coding RNAs demonstrating oncogenic and/or tumor suppressor properties.Citation15
To identify transcriptional changes occurring during invasion, we have performed a comparative microarray analysis of coding and non-coding genes between MCF10A.B2 acini overexpressing p130Cas with or without activation of ErbB2. We have found that genes involved in biological processes like amino acid synthesis (CBS, PHGDH), cell motility and migration (ITPKA, PRDM1), and angiogenesis (HEY1) are upregulated in invasive p130Cas-overexpressing and ErbB2-activated mammary acini, while genes implicated in inflammatory diseases (S100A7 and SAA1) are downregulated. In parallel, we have also identified a microRNA invasive signature in which miR-200b, miR-222, miR-221, miR-210, and miR-424 result to be upregulated, and miR-27a, miR-27b, and miR-23b are downregulated. Interestingly, we have also found that the target genes of miR-27a, miR-27b, and miR-23b are differentially expressed in the invasive signature.
Our results provide new insights on the genetic program underlying p130Cas/ErbB2-dependent invasion that may be useful for prognostic and therapeutic purposes. Indeed, this invasive signature may serve as a prognostic tool to discriminate between non-invasive and invasive ErbB2-positive tumors.
Results
Whole-genome expression analysis reveals two invasive p130Cas/ErbB2-dependent signatures
It has previously been shown that activation of ErbB2 in MCF10A.B2 cells treated with AP1510 (B2) leads to the formation of multiacinar structures that resemble the DCIS condition of human breast cancer.Citation13 Similarly, the overexpression of p130Cas in MCF10A.B2 cells (Cas) induces per se multi acini formation.Citation14 Remarkably, the activation of ErbB2 in Cas cells (Cas/B2) induces invasion into the surrounding extracellular matrix in about 50–60% of the acinar structuresCitation12 ().
Figure 1. Invasive signature of Cas/B2 vs. B2 acini. MCF10A.B2 cells transduced or not with p130Cas-GFP were plated on a matrix generated by mixing Matrigel and collagen I in a 1:1 ratio and stimulated with vehicle or 1 micromolar AP1510 on days 12, 15 and 18. (A) Representative images of day 19 multiacinar structures resulting from ErbB2 activation with AP1510 (B2) or p130Cas overexpression (Cas) and of invasive structures derived from concomitant ErbB2 activation with AP1510 and p130Cas overexpression (Cas/B2). Scale bars, 100 μm. (B) Heat map representing the profile of 91 genes differentially expressed in invasive (Cas/B2) vs. non-invasive (B2) acini. A color-coded scale for the normalized expression values has been used: red represents upregulation and green downregulation of Cas/B2 vs. B2. Arrows indicate the validated genes.
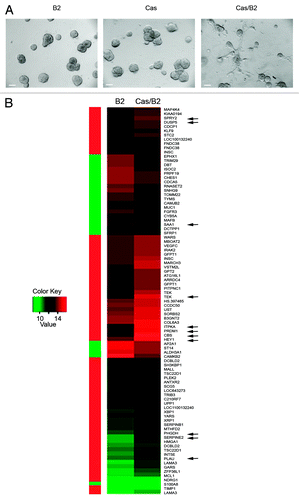
In order to define the gene expression pattern specifically associated with invasiveness of ErbB2-activated acini in presence of p130Cas overexpression, we performed protein-coding microarray analyses (Illumina, HumanHT-12/V3) on high-quality RNA from invasive acini (Cas/B2) and non-invasive (Cas or B2) multiacinar structures as illustrated in . According to our data, two invasive signatures can be identified. The gene profile obtained from the comparison of Cas/B2 vs. B2 acini revealed a signature of 91 differently expressed genes, among which 67 were upregulated and 24 downregulated (; Table S1). On the other hand, the comparison of Cas/B2 vs. Cas acinar structures revealed a signature of 232 genes, among which 160 were upregulated and 72 downregulated (Fig. S1 and Table S2). These findings support that deregulation of specific gene subsets may contribute to the invasive phenotype observed in Cas/B2 transformed cells.
Validation of microarray data
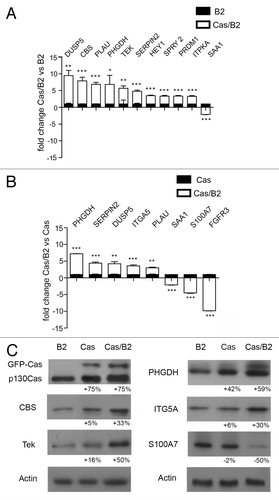
Among the genes differentially expressed in Cas/B2 vs. Cas or B2 acini, we selected some genes for validation by qRT-PCR analysis with a fold change of at least ± 1.5 (Tables S1 and S2). As shown in , among the most upregulated genes in the Cas/B2 vs. B2, we validated DUSP5, CBS, PLAU, PHGDH, TEK, SERPIN2, HEY1, SPRY2, PRDM1, and ITPKA. Instead, among the downregulated genes, we validated SAA1. In the Cas/B2 vs. Cas comparison, PHGDH, SERPIN2, DUSP5, ITGA5, and PLAU resulted as validated among the upregulated genes, while SAA1, S100A7, and FGFR3 were among the downregulated components (). Some of these genes were also validated at protein levels, consistent with mRNA modulations, as shown in . Some of the validated genes were more strongly modulated (up or down) in Cas/B2 invasive acini than in Cas non-invasive acini, suggesting that p130Cas and ErbB2 cooperate in a synergistic manner. Differential expression of some of the genes associated with cell invasiveness was also evaluated in 2D growth conditions and, as shown in Figure S2; in most cases the expression of these genes is modified accordingly. However, CBS, one of the most upregulated genes in Cas/B2 vs. B2 signature was not differentially expressed in 2D culture conditions, suggesting that 3D cultures can unravel gene expression changes relevant for invasion that are lost in 2D cultures.
Figure 2. Whole-genome microarray analysis validation. Gene validation of microarray data by qRT-PCR performed in triplicate on three different RNA preparations from Cas/B2 vs. B2 acini (A) and from Cas/B2 vs. Cas acini (B). Microarray analysis and qRT-PCR fold changes are shown for each validated gene as average values. Bars represent ± standard deviations. The 18S housekeeping gene was used as an internal control to normalize the data (A and B). (C) Total protein cell extracts from B2, Cas and Cas/B2 acini grown in 3D were probed with the antibodies against the indicated proteins. Actin is provided as loading control. Protein modulations were calculated relative to B2 levels, normalized on actin loading controls and expressed as percentages.
These results support the reliability of our microarray analyses for the determination of gene deregulation during invasion.
Functions of the genes belonging to the invasive signatures
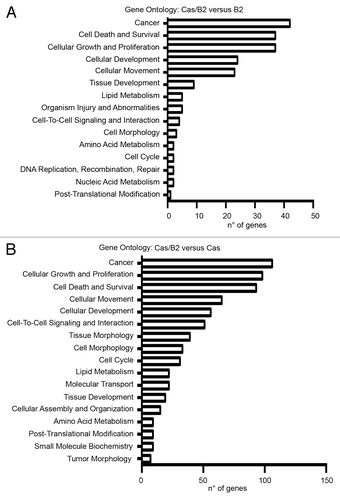
To determine whether the differentially expressed genes in invasive vs. non-invasive 3D cultured cells shared a particular biological function, we conducted a gene ontology (GO) analysis. Notably, our analyses revealed that a significant portion of the over-represented GO annotations points to biological processes linked to tumor progression. As shown in , the main GO categories for over-represented genes in both Cas/B2 compared with B2 or to Cas were “cancer, cell death and survival, cell growth and proliferation, cellular development and movement”, with a P value of <0.001. Overall this analysis highlighted that the most represented categories are those linked to transformation and invasion.
Figure 3. Gene ontology analysis. Gene ontology (GO) analysis of genes differentially expressed in Cas/B2 compared with B2 acini (A) and in Cas/B2 compared with Cas acini (B). The bars indicate the number of over-represented differentially expressed genes. P < 0.001 was used as a threshold to select significant GO categories.
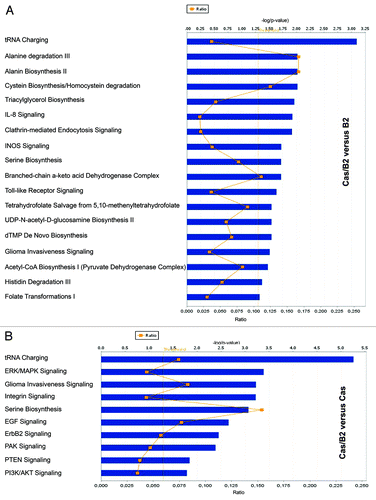
To further evaluate the functional pathways in which the identified genes are involved in p130Cas/ErbB2-dependent invasion, we used the Ingenuity Pathway Analysis (IPA) system. One main molecular network was identified for Cas/B2 vs. B2 gene signature (). This network includes 35 genes associated with “cellular growth and proliferation and cellular movement”, such as tissue inhibitor of metalloproteinases (Timp1), the serine/threonine kinase Mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4), the fibroblast growth factor receptor 3 (FGFR3), PR domain zinc finger protein 1 (PRDM1), Erk1/2, and Hairy/enhancer of split related with YRPW motif 1 (HEY1). In parallel, as shown in , we were able to identify a “cell movement and cancer-related” network for the Cas/B2 vs. Cas gene signature. Although these two networks share some similarities, most of the genes are not overlapping, indicating that the two invasive signatures might result from the integration of differentially activated signaling pathways. This is in line with the pathway analysis shown in , in which it is evident that only some common signaling exist between the Cas/B2 vs. B2 or vs. Cas signatures. Specifically, the most representative signaling in the Cas/B2 vs. B2 signature were those related to amino acid synthesis and degradation, invasiveness, and glucose metabolism. In parallel, the Cas/B2 vs. Cas signature was characterized by a higher number of predicted signaling pathways involved in amino acid transport and metabolism, proliferation, survival, adhesion, and invasion. Interestingly, when we evaluated the correlation of our 91 (Cas/B2 vs. B2) and 232 (Cas/B2 vs. Cas) differentially expressed genes with patient survival in different human breast cancer data sets, we found that in Miller data setCitation16 both up- and downregulated genes of the 91 signature correlated with worse survival (Fig. S3A and B). The same correlation with survival was also observed for the upregulated genes of the 91-gene signature with the Wang data setCitation17 (Fig. S3C). In addition, the entire 232-gene signature (up- and downregulated genes together) showed a significant correlation with survival in the Sotiriou data setCitation18 (Fig. S3D), suggesting the relevance of our data in various large patient cohorts.
Figure 4. Ingenuity Systems Analysis of modulated genes upon invasive conditions. (A) Functional network connecting the 91 differentially expressed invasive genes identified in Cas/B2 vs. B2 acini involved in “cellular growth and proliferation, cellular movement, and cancer” from analyses carried on with Ingenuity™ Pathway Analysis. Score = 50. (B) Functional network connecting the 232 differentially expressed invasive genes identified in Cas/B2 vs. Cas acini involved in “cellular movement, skeletal and muscular system development and function, cancer”. Score = 35. Gene products are represented as nodes and biological relationships between two nodes as a line. Continuous lines indicate direct interactions, while dashed lines represent indirect connections. The green and red symbols represent down- and upregulation respectively, while the white symbols indicate genes absent in the data set but related with the data set genes (A and B).
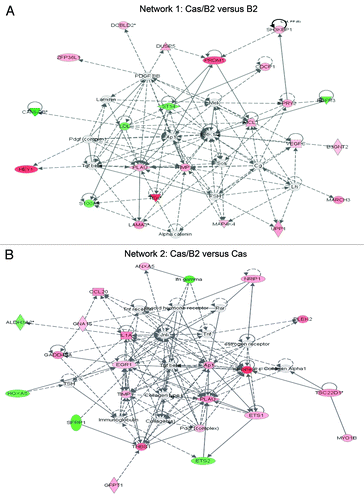
Figure 5. Most representative signaling pathways associated with the two invasive signatures. (A) A total of 91 genes were used to generate functional pathways employing the Ingenuity Pathway Analysis System involved in Cas/B2 vs. B2 invasive signature. (B) A total of 232 genes were used, as in (A), for the identification of the most representative signaling pathways characterizing the Cas/B2 vs. Cas invasive signature.
Modulation of CBS activity and of S100A7 and PHGDH expression levels is sufficient to block p130Cas-dependent invasion of ErbB2 transformed acini
In order to investigate the relevance of the identified invasive signatures, we performed gain- and loss-of-function studies. As shown in , when mammary acini grown in 3D were treated with a chemical inhibitor for CBS (Hydroxylamine [Hydr.]), one of the most upregulated genes in the Cas/B2 vs. B2 signature, (Table S1), the invasive phenotype was abrogated in Cas/B2 acini, while no effects were observed in B2 cells (, panels e and f). Conversely, CBS activation, induced by treatment with S-adenosyl methionine (AdoMet), promoted invasion of B2 acini but was not able to induce invasion of Cas acini in absence of ErbB2 activation (, panels g and h). These results indicate that CBS hyperactivation contributes to the invasive phenotype in an ErbB2-dependent manner. Since we have previously demonstrated that Erk1/2 MAPKs and PI3K/Akt activation are both involved in p130Cas-dependent invasion of ErbB2 transformed acini,Citation13 and CBS was reported to induce Akt and Erk1/2 MAPKs activation in colon cells,Citation19 we tested whether the phosphorylation levels of MAPKs and Akt were affected by treatments with CBS inhibitor and activator. Both Akt and Erk1/2 MAPKs activation were decreased upon treatment with Hydroxylamine and increased in the presence of AdoMet (), indicating that Akt and Erk1/2 MAPKs are implicated in CBS-dependent invasion.
Figure 6. CBS, S100A7, and PHGDH are crucial regulators of Cas/B2 invasion. (A) Representative phase contrast images taken at day 19 of acinar structures formed by unmodified, untreated mock cells (control) and by B2 or Cas or Cas/B2 cells plated in 3D and left untreated or treated with the CBS inhibitor Hydroxylamine (Hydr.) or with the CBS activator S-adenosyl methionine (AdoMet) (panels a-h) as described in the Materials and Methods section. Scale bars, 100 μm. On day 19, acinar structures were recovered and lysed. (B) Total protein cell extracts were analyzed by immunoblot and probed with antibodies to pAkt S473, Akt, pErk1/2 Thr202/Tyr204 and Erk1/2. (C) B2 or Cas or Cas/B2 cells were transduced or not with lentiviral vectors carrying S100A7 cDNA. Total cell extracts showing the levels of S100A7 overexpression (upper panel) and PHGDH downregulation (lower panel) are shown. Actin is provided as loading control. Protein modulations were calculated relative to B2 levels, normalized on actin loading controls and expressed as percentages. (D) B2 and Cas cells overexpressing S100A7 or downregulated for PHGDH were subjected to Transwell invasion assays. After 48 h, cells were fixed and stained. Quantification of cells that invaded through the Matrigel/Collagen matrix from two experiments performed in triplicate and statistical analysis were done as described in Materials and Methods (***p < 0.0001).
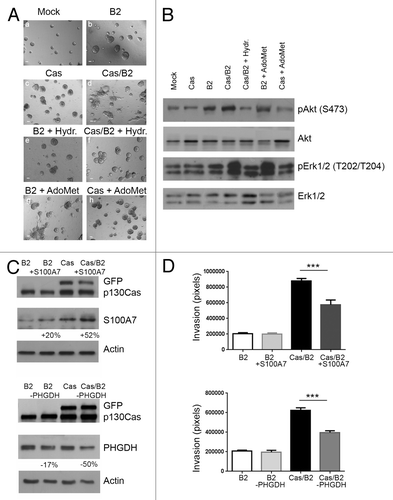
In parallel, Transwell invasion assays performed through a layer of Matrigel/collagen-coated membranes indicated that the overexpression of S100A7, one of the most downregulated genes in the Cas/ErbB vs. Cas signature, was sufficient to inhibit cell invasion in p130Cas overexpressing ErbB2 transformed acini (). In addition, lowering the expression levels of PHGDH, which is upregulated in Cas/B2 vs. B2 or Cas signatures, impairs ErbB2-dependent cell invasion of p130Cas overexpressing acini ().
These data indicate that alteration of S100A7 and PHGDH expression induced by p130Cas are instrumental to induce invasiveness of ErbB2-trasformed acini.
miRNA expression analysis revealed a p130Cas/ErbB2-dependent signature
Increasing evidence points out miRNAs as master regulators of gene expression in many biological and pathological processes, including mammary gland development and breast cancer.Citation20 Therefore, our aim was to evaluate the contribution of specific miRNAs in the acquirement of invasive properties of p130Cas overexpressing cells upon ErbB2 activation. For this purpose, RNA from B2 and Cas/B2was used to hybridize a human microRNA microarray, and 40 differentially expressed miRNAs (20 up- and 20 downregulated) were identified, considering a false discovery rate (FDR) = 8.9% (; Table S3). In order to validate these results, we performed qRT-PCR expression analysis for the differentially expressed microRNAs miR-210, miR-222, miR-424, miR-221, miR-23b, miR-27b, and miR-455-3p. As shown in , miRNA expression patterns were largely consistent between the microarray and the qRT-PCR data.
Figure 7. microRNA expression profiles and target validation in invasive Cas/B2 vs. non-invasive B2 acini. (A) Heat map representing the 40 differentially expressed miRNAs, identified by SAM two class paired analysis (FDR = 8.9%), between invasive Cas/B2 (right column) compared with non-invasive B2 (left column) acini. Each row shows the average expression values (n = 5) referring to individual miRNAs. A color-coded scale for the normalized expression values has been used: red and green represent high and low expression levels, respectively. The expression level of each miRNA was calculated as the Log2 (expression value) and the list of these expression values is provided in the Table S3. (B) MicroRNA expression was analyzed by qRT-PCR in triplicate for 7 miRNAs on 5 different RNA preparations from invasive Cas/B2 and non-invasive B2 acini. U6 small nucleolar RNA expression was used as internal control. Bar represent ± standard deviations. (C) B2 or Cas or Cas/B2 cells overexpressing pre-control or pre-miR-210 were subjected to Transwell invasion assays. After 48 h, cells were fixed and stained. Quantification of cells that invaded through the Matrigel/Collagen matrix from two experiments performed in triplicate and statistical analysis were performed as described in Materials and Methods (**p < 0.001), (***p < 0.0001). (D) Levels of miR-210 upregulation obtained after transfection of cells used in (C). (E) Schematic diagram illustrating the strategy for finding miRNA target genes in the invasive 91-gene coding signature.
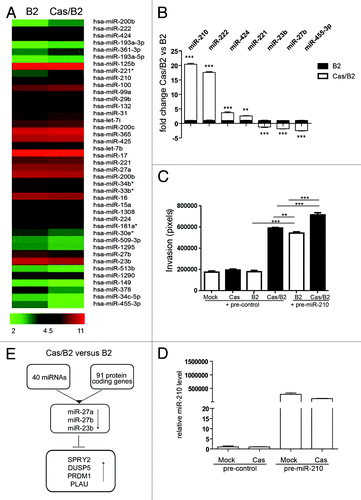
Since miR-210 has been recently described to be involved in tumor proliferation, invasion, and poor clinical outcome in breast cancer,Citation21,Citation22 to gain functional insights on the role of this miRNA in mediating invasion, Mock, Cas, B2 or Cas/B2 cells were transfected with pre-control or pre-miR-210 and used in a matrigel/collagen Transwell invasion assay (). Transfected cells were treated with AP1510 (ErbB2 activator) immediately before seeding the cells. Upregulation of miR-210 is shown in . As shown in , as expected, invasion was promoted only in Cas/B2 cells in control conditions. Interestingly, the transfection of pre-miR-210 was sufficient to promote invasion in B2 cells and to further increase invasion in Cas/B2 cells. These results indicate that miR-210 is indeed playing a critical role in ErbB2 transformed cell invasion and that the concomitant overexpression of p130Cas and miR-210 in ErbB2 transformed cells boosts their invasive behavior.
The identification of miRNA targets is crucial to understand the biological role of miRNAs. Since mRNA destabilization is one of miRNA-mediated gene repression mechanisms, we used the TargetScan (v. 5.1) algorithm to identify putative targets of the differentially expressed miRNAs in Cas/B2 vs. B2 acini (; Table S3) and looked for overlaps with the Cas/B2 vs. B2 signature (; Table S1). Gene targets of the downregulated miRNAs, such as miR-27a, miR-27b, and miR-23b, were identified () and are consistently upregulated (Table S4).
Discussion
Cancer relapse and resultant metastatic spread to distant sites are still the leading cause of cancer-related deaths in the world.Citation23 Several studies have been designed to assess the genetic alterations associated with breast cancer recurrence and progression.Citation24 Despite these studies, a detailed molecular characterization of breast cancer progression is still missing. In this study, by inducing ErbB2 activation in the presence or in the absence of p130Cas overexpression, we provided an in vitro cell model for the understanding of the genetic changes that are associated with the shift from a non-invasive to an invasive mammary tumor. Specifically, by comparing ErbB2 and Cas multiacinar non-invasive cells with invasive Cas/B2 cells in 3D, we have identified differentially expressed protein-coding genes and miRNAs that are deregulated during invasion driven by p130Cas in ErbB2-transformed cells.
It is now well established that, in order to acquire an invasive phenotype, mammary transformed cells require the upregulation of genes involved in epithelial–mesenchymal transition (EMT), migration,Citation25 and small GTPases activation.Citation26,Citation27 Accordingly, among the differentially expressed genes in invasive vs. non-invasive acini, we found a significant upregulation of genes previously implicated in these cellular processes. Among them, PRDM1 that encodes the transcriptional repressor Blimp-1Citation28 was recently described as a potent repressor of ER-α synthesis, which is induced by activation of the NFκB member RelB, leading to a migratory phenotype.Citation29 Moreover, Blimp1 is emerging as a crucial downstream effector of RelB/Bcl-2/Ras pathway that promotes EMT signature and breast cancer cell migration.Citation30 In addition, we observed increased expression of ITPKA and HEY1 genes previously described to promote cell motility, EMT, and migration. Indeed, HEY1 is one of the Notch target genes highly expressed in a large percentage of breast cancer cells during hypoxia and is involved in the initiation of EMT and in promotion of metastasis.Citation31 A correlation between ErbB2 and ITPKA during tumor progression was already reported.Citation32,Citation33 Moreover, overexpression of ITPKA in lung carcinoma cells induces the formation of actin-based cell protrusions and increases migration, confirming that ectopic expression of ITPKA in highly invasive tumor cells is critical for invasiveness and migration.Citation34 It has been recently reported that the MTHFD2 gene, which we found strongly upregulated in Cas vs. B2 invasive condition, is a crucial regulator of breast cancer migration and invasion into extracellular matrix,Citation35 further confirming the validity of the invasive signature. IPA analysis of the Cas/B2 invasive signatures highlights signaling pathways related to amino acid transport and metabolism thus providing additional strength to the hypothesis that changes in metabolism frequently may play a crucial role in aggressive cancer cells. Some evidenceCitation36 points out that increased glycolysis allows the diversion of glycolytic intermediates into various biosynthetic pathways, including those generating nucleosides and amino acids; this facilitates, in turn, the biosynthesis of the macromolecules and organelles required for assembling new cells. Consistently, our data indicate a strong upregulation of PHGDH and CBS genes that are both involved in amino acid biosynthesis. PHGDH is an enzyme of the serine biosynthesis pathway and has also been implicated in glucose uptake required for tumorigenesis.Citation37 It has been found overexpressed in a subset of melanoma and breast cancers, mainly in estrogen receptor-negative and basal-like breast tumors.Citation38 Its upregulation was already demonstrated in MCF10A cells overexpressing ErbB2, resulting in glucose-derived serine production.Citation39 Our data further strengthen the important implications of PHGDH in ErbB2 transformed cells, as downregulation of PHGDH expression is sufficient to halt invasion of Cas/B2 cells. The CBS gene is the key regulatory enzyme for the trans-sulfuration pathway and catalyzes the condensation of homocysteine with serine to create cystathionine, the precursor of cysteine,Citation40 but its role in breast cancer is still unknown. Interestingly, our data indicate that levels of CBS activity seem to be crucial for cell invasiveness. CBS activation promotes Cas/B2 invasion, whereas its inhibition blocks invasion. These results unravel, for the first time, the potential role of CBS in the modulation of ErbB2-tranformed mammary cells. The acquirement of invasiveness implies downregulation of specific gene subsets as well. Interestingly, S100A7 and SAA1 are found to be downregulated during p130Cas-dependent invasion. S100A7 was identified as one of the most abundant transcripts in high-grade ductal carcinoma in situ (DCIS).Citation41 Previous reports showed that S100A7 expression was upregulated in a mammary DCIS model, while its downregulation has been observed during the transition from DCIS to invasive cancer.Citation42 Moreover, it was proposed that the silencing of S100A7 is required for both initiation of tumorigenesis and invasiveness in breast cancer cells.Citation43 Noteworthy, our data indicate that re-establishment of S100A7 expression levels by lentiviral infection is sufficient in an in vitro invasion assay to impair cell invasion, further sustaining the critical role of this protein in the regulation of breast cancer invasion. SAA proteins were found elevated in different malignancies, such as kidney, colon, and prostate cancers as well as in leukemias and lymphomas.Citation44 Furthermore, SAA1 is a well-known downstream target of C/EBP β and Δ.Citation45 SAA1 has been implicated in the Stat3-dependent transcription regulatory network controlling metastatic progression.Citation46
Since p130Cas is a versatile adaptor molecule, changes of its expression may be sufficient to deregulate simultaneously a number of signaling pathways, thereby causing extensive gene expression alterations required for cell invasion. Indeed, IPA analysis suggests that p130Cas-dependent invasion of ErbB2 transformed cells is associated with activation of multiple signaling pathways related to a variety of cell functions. As a consequence, the invasive process may be the result of signaling pathway integration into a complex network converging on MAPKs activity. Our data indeed provide evidence of additional cell functions whereby p130Cas exerts its pro-invasive role. Beside the well-established ability of p130Cas to regulate cytoskeleton re-organization during cell invasion,Citation11 here, for the first time, we describe that high levels of p130Cas impact on the transcription of genes implicated in cell migration as well as in tumor cell metabolism. In addition, it is worth noting that the comparison of the differentially expressed coding-genes with different human breast cancers provides a correlation with patient survival, suggesting a prognostic value of the here-identified p130Cas/ErbB2-dependent signatures.
It is now emerging that miRNAs have an important role in cancer predisposition, initiation, and progression. In the last years, accumulating evidence has indicated that miRNAs influence cancer cell adhesion, migration, invasion, motility, and angiogenesis.Citation47,Citation48 Understanding the detailed molecular mechanisms through which miRNAs lead to cancer progression can be useful to identify new markers for breast cancer progression and new therapeutic targets.
Among the differential expressed miRNAs in invasive Cas/B2 vs. non-invasive B2 acini, we found that miR-200b, miR-210, miR-222, miR-424, miR-221 are upregulated, while miR-455-3p, miR-23b, and miR-27b are downregulated.
Upregulation of miR-200b, miR-222, and miR-221 has been recently correlated with invasive behavior of breast cancer cells,Citation49,Citation50 while miR-424 has been described to play a role in pro-angiogenesis.Citation51
Interestingly, miR-210 was recently associated with tumor proliferation, invasion, and poor clinical outcome in breast cancer,Citation21 and miR-210 is upregulated in the transition from in situ to invasive breast carcinoma.Citation22 Our functional data further confirm the role of miR-210 in breast cancer invasion, indicating that its upregulation is sufficient to promote the transition from non-invasive to invasive of ErbB2 transformed acini.
Conversely, downregulation of miR-23b is known to inhibit both migration and invasion and to induce apoptosis of colon cancer cells.Citation52 In addition, in hepatocellular carcinoma, low expression of miR-23b has been shown to induce the urokinase-type plasminogen activator (uPA), which, by promoting extracellular matrix degradation, favors cell invasion.Citation53 In different breast cancer subsets miR-27b downregulation has been associated with upregulation of Cytochrome P450, a drug-metabolizing enzyme.Citation54 However, so far, no evidence on its implications in cancer progression has been described.
No data are available in literature on the role of miR-27a in cancer invasion, although in breast cancer cell lines the overexpression of this miRNA induces cell cycle progression.Citation54,Citation55 Downregulation of genes that trigger proliferation has been already observed in invasive cells, indicating that a reduced proliferation is required for cells to turn on the invasive program.Citation56
An anti-correlation between miRNAs and putative target genes was found for the downregulated miR-23b, miR-27a, and miR-27b. Specifically, PRDM1, DUSP5, SPROUTY2, and PLAU expression levels are increased in invasive compared with non-invasive acini.
In conclusion, these findings show that p130Cas is able to induce a specific invasive signature resulting from deregulation of protein coding genes and miRNAs. This signature may be useful to discriminate between invasive and non-invasive ErbB2-positive tumors with high levels of p130Cas, and it might be helpful for the development of new therapeutic drugs for the treatment of aggressive breast cancer.
Materials and Methods
Antibodies and reagents
Mouse monoclonal antibodies to p130Cas and to ITGA5 were produced at the MBC, University of Torino. Anti-S100A7 monoclonal Abs were purchased from Imgenex, anti-PHGDH monoclonal Abs from Abcam, anti-Tek and anti-actin policlonal Abs from Santa Cruz, anti-CBS monoclonal Abs were from GeneTex, anti-phospho-Erk1/2 (T202/Y204), anti-phospho-Akt (S473), anti-Akt, anti-Erk1/2 antibodies were from Cell Signaling. CBS inhibitor (Hydroxylamine) and CBS activator (S-Adenosyl methionine, Adomet) were purchased from Sigma. Matrigel and collagen I were purchased from BD Transduction Laboratories (Franklin Lakes). Secondary antibodies conjugated with peroxidase were purchased from GE Healthcare. miR precursors: Pre-miR™ miRNA Precursor Negative Control #1 and Pre-miR™ miRNA Precursor Hsa-miR-210 (PM10516), were all from Applied Biosystems.
Cell lines and cultures
MCF10A.B2 engineered cells were kindly provided by Dr Muthuswamy (Cold Spring Harbor Lab) and described in Muthuswamy et al., 2001.Citation15 MCF10A.B2 cells were cultivated in DMEM/F12 (Gibco, BRL) supplemented with 5% horse serum, 20 ng/ml EGF, 10 μg/ml insulin, 1 ng/ml cholera toxin, 100 μg/ml hydrocortisone, 50 U/ml penicillin, and 50 μg/ml streptomycin. To generate 10A.B2Mock and 10A.B2Cas cells, we transduced MCF10A.B2 with pBabe retroviral empty vector or carrying p130Cas cDNA fused with GFP as described in Cabodi et al., 2011 ().Citation12
Table 1. Cell lines and cultures
Three-dimensional morphogenetic assay
Three-dimensional morphogenetic assays were conducted as previously described in http://muthuswamylab.cshl.edu/ml_protocols.html and in.Citation57,Citation58 Briefly, MCF10A.B2 cells were seeded in a Matrigel:Collagen mixture (Growth factor reduced Matrigel, #354230 BD; Bovine Collagen Type I #354231, BD). Cells were left to grown for 12 d, changing the culture medium every 4 d. At 12 d, acini were formed in 3D and left unstimulated or stimulated every 3 d with 1 micromolar of AP1510 (Ariad). Phase images of day 18 acini were collected by using Apotome Zeiss microscopy at 10× magnitude. In the experiments performed with CBS inhibitor, acini were pre-treated for 1 d with the chemical compounds (0.5 mM Hydroxylamine; 100 micromolar S-Adenosyl methionine, Adomet) and stimulated at day 12 with AP1510. CBS activator AdoMet was added at 12 d concomitantly with AP1510.
Transient transfections
To obtain transient pre-control or pre-miR-210 expression, Mock, Cas, B2, or Cas/B2 cells were plated at 50% confluency and immediately transfected using HyPerFect Transfection Reagent (QIAGEN) reagent, with 75 nM pre-miR.
Lentiviral constructs
For S100A7 overexpression, human S100A7 cDNA (Addgene) was cloned into pCCL lentiviral vector, and viral particles production was performed as previously described.Citation12 PHGDH shRNA lentiviral particles were purchased from Sigma.
Transwell invasion assay
Experiments were conducted in agreement with previous protocols (http://www.lbl.gov/lifesciences/BissellLab/labprotocols/invasionassay.htm). Briefly, 24-transwell chambers (Corning Costar) were coated with 50 microliters of 1:1 mix matrigel plus collagen. 105 infected or transfected cells in 100 μl of medium without FBS were seeded into the upper chamber. Five hundred microliters of DMEM-F12 with 5% horse serum plus AP1510 or ethanol for control, were added into the lower chamber as chemotactic stimulus. After 48 h, the migrated cells on the lower side of the membrane were fixed in 2.5% glutaraldehyde, stained with 0.1% crystal violet and photographed using an Olympus IX70 microscope. Invasion was evaluated by measuring the area occupied by migrated cells using the ImageJ software (http://rsbweb.nih.gov/ij/).
Protein extracts preparation from 3D cultures and western blotting analysis
3D acini were released from Matrigel:Collagen using BD cell recovery solution (BD Biosciences). Briefly, cell recovery solution was added to cells embedded in Matrigel:collagen. The cell mixture was recovered and placed on ice for 30 min to dissolve the Matrigel. Cells were centrifuged for 3 min at 4 °C and lysed with RIPA buffer (50 mM TRIS-HCl pH 7.4, 150 mM NaCl; 1% Triton X-100, and protease and phosphatase inhibitors). Total cell lysates were separated by SDS-PAGE, transferred to nitrocellulose and immunoblotted overnight with primary antibodies at 4 °C. Blots were incubated with mouse or rabbit horseradish-peroxidase conjugated secondary antibodies for 1h at room temperature. ECL (Euroclone) was used to detect chemoluminescent signals. Protein band intensities were determined using the ImageJ software developed by the NIH.
RNA isolation and analysis from 3D cultures
Total RNA from Mock, B2, Cas, and Cas/B2 cells was isolated by using Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. RNA quantification was performed using the NanoDrop-1000 spectrophotometer (Nanodrop). Total RNA integrity in each sample was assessed by capillary electrophoresis using the Agilent Bioanalyzer 2100 with the RNA 6000 Nano and the Small RNA Nano LabChips respectively (Agilent Technologies). One microgram of DNase-treated (DNA-free™ kit, Ambion) total RNA was reverse transcribed using RETROscript™ reagents (Ambion). RNA was heat-denatured for 3′ at 85 °C and the reaction was incubated at 42 °C for 1h10m at 92 °C. Quantitative Real Time PCR (qRT-PCR) reactions were performed in 96 well plates using SYBRGreen® Master Mix (Applied Biosystems), specific primers and 10 ng total RNA converted into cDNA in 10 microliter final volume. Fluorescence was measured using an ABI Prism® 7300 (Applied Biosystems) detection system according to the manufacturer’s instructions. Primers were purchased from QIAGEN (QuantiTect® Primer Assays). Relative quantifications to control cells were performed: first, each Ct value was corrected for the Ctr of the reference gene (GAPDH), and then the Ct of each sample was subtracted from the Ct of control cells (Ct0). The relative amount of template (Q) was therefore calculated as: Q = 2 − (Ct − Ct r) − (Ct 0 − Ct 0r). All samples were run in triplicates and mean and standard deviation calculated as described in Bookout et al.Citation58
Whole genome expression profiling
Microarray analysis of gene expression in response to p130Cas overexpression and ErbB2 activation in Cas/B2 vs. B2 and Cas cells was performed using the Illumina BeadChip system (Illumina, Inc). We used 500 ng of total RNA to obtain labeled, amplified cRNA for each sample to hybridize the Illumina HumanHT-12_V4 BeadChips according to manufacturer’s instructions (Illumina, Inc). Arrays were scanned with an IlluminaBeadArray Reader confocal scanner and data processed and rank invariant normalized using GenomeStudio software (Illumina, Inc). Normalized data were Log2-transformed and analyzed with Excel (Microsoft) to assess differential expression between the Cas/B2 and Cas or B2 control clones based on three RNA preparations from each clone. Probes were first filtered by their “detection” P value, as calculated by GenomeStudio, which had to be 0.001 or lower in each of the three samples belonging to the group with the highest average expression. This filter, which also included a control group not overexpressing p130Cas or ErbB2, was passed by 12 235 probes. To be defined as differential between 2 groups, a probe signal had to display significance in all the following three basic tests: (1) fold-change between the means of the 2 groups greater than a threshold (1.5 for the Cas/B2 vs. Cas and 1.7 for the Cas/B2 vs. B2 comparisons); (2) independent samples, two-way t test P value lower than 0.005; (3) signal-to-noise ratio greater than 3 (positive or negative), according to the formula described by Golub and colleagues.Citation59 Montecarlo simulation with sample permutation analysis confirmed that under the conditions described above the FDR was well below 5%. Raw data are available on the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE43907.
miRNAs expression profiling
miRNAs expression profiles were performed using the “Human microRNA Microarray kit (V3)” (Agilent Technologies), which allows the detection of 866 known human (miRBase v.12) and 89 human viral miRs. Each slide contains eight individual microarrays, with ~15 000 features each, including 48 negative controls, used to estimate fluorescence background and background variance. Each miRNA was targeted by 16–20 array-probes of different sizes. Total RNA (200 ng) was labeled with pCp Cy3, according to the Agilent protocol, and unincorporated dyes were removed with MicroBioSpin6 columns (BioRad). Probes were hybridized at 55 °C for 22 h using the Agilent hybridization oven that is suited for bubble-mixing and microarray hybridization processes. Slides were washed by Agilent Gene expression wash buffer 1 and 2 and scanned using an Agilent microarray scanner (model G2565CA) at 100% and 5% sensitivity settings. Raw data are available on the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE42949.
Statistical analysis of miRNA and gene expression data
Inter-array normalization of expression levels was performed with cyclic Lowess for miRNA experiments to correct possible experimental distortions.Citation60 Normalization function was applied to expression data of all experiments and then values of spot replicates within arrays were averaged.
For miRNAs, Feature Extraction Software was used to obtain spot quality measures in order to evaluate the quality and the reliability of the hybridization. In particular, flag ‘glsFound’ (set to 1 if the spot has an intensity value significantly different from the local background, 0 otherwise) was used to filter out unreliable probes: flag equal to 0 will be noted as “null”. So, in order to make more robust and unbiased statistical analyses, probes with a high proportion of NA values were removed from the data set. Forty percent of “null” was used as threshold in the filtering process, obtaining a total of 175 available human miRs. Principal Component Analysis (PCA), cluster analysis and profile similarity searches were performed with Multi Experiment Viewer, version 4.6.2 (tMev), of the TM4 Microarray Software Suite.Citation61 All heat maps were obtained by tMeV software using an unsupervised two-dimensional hierarchical clustering approach with average linkage method and Pearson correlation. The identification of differentially expressed miRNAs was performed with two class paired Significance Analysis of Microarray (SAM) programCitation62 with default settings. The expression level of each miRNA was calculated as the Log2(Cas/B2/B2).
qRT-PCR for coding genes and miRNAs detection
The isolated total RNA samples with A260/A280 ratios between 1.8 and 2.0, was reverse transcribed using High Capacity cDNA Reverse Transcription kit (Applied Biosystems). RNA were incubated for 10 min at 25 °C, then at 37 °C for 2 h and 5 s at 85 °C. The obtained cDNA was diluted 1:5 in RNAase free water. Quantitative RT-PCR was performed on cDNA using the 7900HT Fast Real-Time PCR System (Applied Biosystems) with SYBRGreen® Master Mix (Applied Biosystems). Primers for SAA1, PHGDH, DUSP5, SPROUTY2, ITGA5, TEK, PLAU, and SERPINE2, were obtained from Qiagen. Primer sequences for CBS, HEY1, PRDM1, ITPKA, S100A7, and FGFR3 were obtained from Sigma. Primer sequences are available upon request. Conditions were as follows: 50 °C for 2 min, 95 °C for 2 min, followed by 45 cycles of 90 °C for 15 s and 60 °C for 30 s. Total volume was of 20 microliters. Quantitative normalization was performed on the expression of endogenous control 18S. qRT-PCRs for microRNA detection were performed with the indicated TaqMan MicroRNA Assays (Applied Biosystems) on 10 ng total RNA, according to the manufacturer’s instructions. For mRNA detection, 1 µg of DNase-treated RNA (DNA-free™ kit, Ambion) was retro-transcribed with RETROscript™ reagents (Ambion), and qRT–PCRs were performed using gene-specific primers, using a 7900HT Fast Real-Time PCR System. Quantitative normalization was performed on the expression of the U6 small nucleolar RNA. The relative expression levels between samples were calculated using the comparative delta CT (threshold cycle number) method (2-ΔΔCT) with a control sample as the reference point.Citation58
Target Prediction and pathways analysis
The TargetScan algorithm, release 5.1Citation63 was used to predict miR targets. The Ingenuity Pathways Knowledge Base (http://www.ingenuity.com) is currently the world's largest database of knowledge on biological networks, with annotation curated by experts. We exploited this database to define the presence of functional associations within the genes detected by microarray analysis and to identify differences between the ontological gene classes that were enriched among differentially expressed genes. This ontological gene classification provides the controlled vocabulary to describe gene and gene product attributes. In addition, we took advantage of this database to search for enrichments of specific pathways among invasive vs. noninvasive conditions. Enrichment significance in pathways analysis is shown as the negative Log10 of the P value. The P value is calculated with the right-tailed Fisher exact test. Ingenuity networks are scored based on the number of network eligible molecules they contain. The network score is based on the hypergeometric distribution and is calculated with the right-tailed Fisher exact test. The score is the negative log of this P value.
Human data set analysis
Two groups of patients were created by Pearson correlation-based hierarchical clustering, using signature expression profiles. Then we evaluated if the cumulative probability of survival was significantly different between the 2 groups by using survival library of R. In Figure S3 we show the Kaplan-Meier curves of different survival probability.
Statistical analyses
Data are presented as mean ± standard deviation (SD) or as mean ± standard error of mean (SEM), as indicated, and two-tailed Student t test was used for comparison, with * = P < 0.05; ** = P < 0.01; *** = P < 0.001 considered to be statistically significant. ns indicates a non-statistically significant P value.
Additional material
Download Zip (663.8 KB)Acknowledgments
This work was supported by AIRC grants IG11346 to SC, IG10104 to DT, IG5975 to PP, and IG11896 to PD, MIUR (FIRB giovani 2008 RBFR08F2FS to SC and FO), the Regione Piemonte POR F.E.S.R.2007/2013 “DRUIDI: Piattaforme Innovative per le Scienze della Vita” to SC and Progetto Sanità Finalizzata Giovani Ricercatori 2009 to SC. G Tornillo and B Bisaro are supported by FIRC fellowships.
Disclosure of potential conflicts of interest
The authors declare no conflicts of interest
Supplemental Materials
Supplemental materials may be found here: http://www.landesbioscience.com/journals/cc/article/25415
References
- Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006; 24:2137 - 50; http://dx.doi.org/10.1200/JCO.2005.05.2308; PMID: 16682732
- Polyak K. Breast cancer: origins and evolution. J Clin Invest 2007; 117:3155 - 63; http://dx.doi.org/10.1172/JCI33295; PMID: 17975657
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244:707 - 12; http://dx.doi.org/10.1126/science.2470152; PMID: 2470152
- Tan M, Yao J, Yu D. Overexpression of the c-erbB-2 gene enhanced intrinsic metastasis potential in human breast cancer cells without increasing their transformation abilities. Cancer Res 1997; 57:1199 - 205; PMID: 9067293
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2002; 2:451 - 61; http://dx.doi.org/10.1016/S1535-6108(02)00212-X; PMID: 12498714
- Feng C, Neumeister V, Ma W, Xu J, Lu L, Bordeaux J, et al. Lin28 regulates HER2 and promotes malignancy through multiple mechanisms. Cell Cycle 2012; 11:2486 - 94; http://dx.doi.org/10.4161/cc.20893; PMID: 22713243
- Raven JF, Williams V, Wang S, Tremblay ML, Muller WJ, Durbin JE, et al. Stat1 is a suppressor of ErbB2/Neu-mediated cellular transformation and mouse mammary gland tumor formation. Cell Cycle 2011; 10:794 - 804; http://dx.doi.org/10.4161/cc.10.5.14956; PMID: 21311224
- Liu E, Thor A, He M, Barcos M, Ljung BM, Benz C. The HER2 (c-erbB-2) oncogene is frequently amplified in in situ carcinomas of the breast. Oncogene 1992; 7:1027 - 32; PMID: 1349163
- Feigin ME, Muthuswamy SK. ErbB receptors and cell polarity: new pathways and paradigms for understanding cell migration and invasion. Exp Cell Res 2009; 315:707 - 16; http://dx.doi.org/10.1016/j.yexcr.2008.10.034; PMID: 19022245
- Rafn B, Kallunki T. A way to invade: a story of ErbB2 and lysosomes. Cell Cycle 2012; 11:2415 - 6; http://dx.doi.org/10.4161/cc.20860; PMID: 22713242
- Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer 2010; 10:858 - 70; http://dx.doi.org/10.1038/nrc2967; PMID: 21102636
- Cabodi S, Tinnirello A, Bisaro B, Tornillo G, del Pilar Camacho-Leal M, Forni G, et al. p130Cas is an essential transducer element in ErbB2 transformation. FASEB J 2010; 24:3796 - 808; http://dx.doi.org/10.1096/fj.10-157347; PMID: 20505116
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol 2001; 3:785 - 92; http://dx.doi.org/10.1038/ncb0901-785; PMID: 11533657
- Tornillo G, Bisaro B, Camacho-Leal MdelP, Galiè M, Provero P, Di Stefano P, et al. p130Cas promotes invasiveness of three-dimensional ErbB2-transformed mammary acinar structures by enhanced activation of mTOR/p70S6K and Rac1. Eur J Cell Biol 2011; 90:237 - 48; http://dx.doi.org/10.1016/j.ejcb.2010.09.002; PMID: 20961652
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065 - 70; http://dx.doi.org/10.1158/0008-5472.CAN-05-1783; PMID: 16103053
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A 2005; 102:13550 - 5; http://dx.doi.org/10.1073/pnas.0506230102; PMID: 16141321
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005; 365:671 - 9; PMID: 15721472
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006; 98:262 - 72; http://dx.doi.org/10.1093/jnci/djj052; PMID: 16478745
- Cai WJ, Wang MJ, Ju LH, Wang C, Zhu YC. Hydrogen sulfide induces human colon cancer cell proliferation: role of Akt, ERK and p21. Cell Biol Int 2010; 34:565 - 72; http://dx.doi.org/10.1042/CBI20090368; PMID: 20184555
- Piao HL, Ma L. Non-coding RNAs as regulators of mammary development and breast cancer. J Mammary Gland Biol Neoplasia 2012; 17:33 - 42; http://dx.doi.org/10.1007/s10911-012-9245-5; PMID: 22350981
- Rothé F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One 2011; 6:e20980; http://dx.doi.org/10.1371/journal.pone.0020980; PMID: 21738599
- Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci U S A 2013; 110:7413 - 7; http://dx.doi.org/10.1073/pnas.1304977110; PMID: 23589849
- Comen EA. Tracking the seed and tending the soil: evolving concepts in metastatic breast cancer. Discov Med 2012; 14:97 - 104; PMID: 22935206
- Njiaju UO, Olopade OI. Genetic determinants of breast cancer risk: a review of current literature and issues pertaining to clinical application. Breast J 2012; 18:436 - 42; http://dx.doi.org/10.1111/j.1524-4741.2012.01274.x; PMID: 22957996
- Berx G, Raspé E, Christofori G, Thiery JP, Sleeman JP. Pre-EMTing metastasis? Recapitulation of morphogenetic processes in cancer. Clin Exp Metastasis 2007; 24:587 - 97; http://dx.doi.org/10.1007/s10585-007-9114-6; PMID: 17978854
- Katz E, Sims AH, Sproul D, Caldwell H, Dixon MJ, Meehan RR, et al. Targeting of Rac GTPases blocks the spread of intact human breast cancer. Oncotarget 2012; 3:608 - 19; PMID: 22689141
- Minn AJ, Bevilacqua E, Yun J, Rosner MR. Identification of novel metastasis suppressor signaling pathways for breast cancer. Cell Cycle 2012; 11:2452 - 7; http://dx.doi.org/10.4161/cc.20624; PMID: 22659842
- Davis MM. Blimp-1 over Budapest. Nat Immunol 2007; 8:445 - 7; http://dx.doi.org/10.1038/ni0507-445; PMID: 17440448
- Wang X, Belguise K, O’Neill CF, Sánchez-Morgan N, Romagnoli M, Eddy SF, et al. RelB NF-kappaB represses estrogen receptor alpha expression via induction of the zinc finger protein Blimp1. Mol Cell Biol 2009; 29:3832 - 44; http://dx.doi.org/10.1128/MCB.00032-09; PMID: 19433448
- Romagnoli M, Belguise K, Yu Z, Wang X, Landesman-Bollag E, Seldin DC, et al. Epithelial-to-mesenchymal transition induced by TGF-β1 is mediated by Blimp-1-dependent repression of BMP-5. Cancer Res 2012; 72:6268 - 78; http://dx.doi.org/10.1158/0008-5472.CAN-12-2270; PMID: 23054396
- Chen J, Imanaka N, Chen J, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer 2010; 102:351 - 60; http://dx.doi.org/10.1038/sj.bjc.6605486; PMID: 20010940
- Windhorst S, Fliegert R, Blechner C, Möllmann K, Hosseini Z, Günther T, et al. Inositol 1,4,5-trisphosphate 3-kinase-A is a new cell motility-promoting protein that increases the metastatic potential of tumor cells by two functional activities. J Biol Chem 2010; 285:5541 - 54; http://dx.doi.org/10.1074/jbc.M109.047050; PMID: 20022963
- Chang L, Schwarzenbach H, Meyer-Staeckling S, Brandt B, Mayr GW, Weitzel JM, et al. Expression Regulation of the Metastasis-Promoting Protein InsP3-Kinase-A in Tumor Cells. Mol Cancer Res 2011; http://dx.doi.org/10.1158/1541-7786.MCR-10-0556; PMID: 21460179
- Windhorst S, Kalinina T, Schmid K, Blechner C, Kriebitzsch N, Hinsch R, et al. Functional role of inositol-1,4,5-trisphosphate-3-kinase-A for motility of malignant transformed cells. Int J Cancer 2011; 129:1300 - 9; http://dx.doi.org/10.1002/ijc.25782; PMID: 21792881
- Lehtinen L, Ketola K, Mäkelä R, Mpindi JP, Viitala M, Kallioniemi O, et al. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget 2013; 4:48 - 63; PMID: 23295955
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev 2008; 18:54 - 61; http://dx.doi.org/10.1016/j.gde.2008.02.003; PMID: 18387799
- Luo J. Cancer’s sweet tooth for serine. Breast Cancer Res 2011; 13:317; http://dx.doi.org/10.1186/bcr2932; PMID: 22189202
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011; 476:346 - 50; http://dx.doi.org/10.1038/nature10350; PMID: 21760589
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 2011; 43:869 - 74; http://dx.doi.org/10.1038/ng.890; PMID: 21804546
- Gupta S, Kruger WD. Cystathionine beta-synthase deficiency causes fat loss in mice. PLoS One 2011; 6:e27598; http://dx.doi.org/10.1371/journal.pone.0027598; PMID: 22096601
- Petersson S, Bylander A, Yhr M, Enerbäck C. S100A7 (Psoriasin), highly expressed in ductal carcinoma in situ (DCIS), is regulated by IFN-gamma in mammary epithelial cells. BMC Cancer 2007; 7:205; http://dx.doi.org/10.1186/1471-2407-7-205; PMID: 17986321
- Krop I, März A, Carlsson H, Li X, Bloushtain-Qimron N, Hu M, et al. A putative role for psoriasin in breast tumor progression. Cancer Res 2005; 65:11326 - 34; http://dx.doi.org/10.1158/0008-5472.CAN-05-1523; PMID: 16357139
- Rhee DK, Park SH, Jang YK. Molecular signatures associated with transformation and progression to breast cancer in the isogenic MCF10 model. Genomics 2008; 92:419 - 28; http://dx.doi.org/10.1016/j.ygeno.2008.08.005; PMID: 18804527
- Malle E, Sodin-Semrl S, Kovacevic A. Serum amyloid A: an acute-phase protein involved in tumour pathogenesis. Cell Mol Life Sci 2009; 66:9 - 26; http://dx.doi.org/10.1007/s00018-008-8321-x; PMID: 18726069
- Ray A, Ray BK. Serum amyloid A gene expression under acute-phase conditions involves participation of inducible C/EBP-beta and C/EBP-delta and their activation by phosphorylation. Mol Cell Biol 1994; 14:4324 - 32; PMID: 8196668
- Ranger JJ, Levy DE, Shahalizadeh S, Hallett M, Muller WJ. Identification of a Stat3-dependent transcription regulatory network involved in metastatic progression. Cancer Res 2009; 69:6823 - 30; http://dx.doi.org/10.1158/0008-5472.CAN-09-1684; PMID: 19690134
- Shi M, Liu D, Duan H, Shen B, Guo N. Metastasis-related miRNAs, active players in breast cancer invasion, and metastasis. Cancer Metastasis Rev 2010; 29:785 - 99; http://dx.doi.org/10.1007/s10555-010-9265-9; PMID: 20938719
- Siragam V, Rutnam ZJ, Yang W, Fang L, Luo L, Yang X, et al. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting activin receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget 2012; 3:1370 - 85; PMID: 23211491
- Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One 2009; 4:e7181; http://dx.doi.org/10.1371/journal.pone.0007181; PMID: 19787069
- Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, et al. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal 2011; 4:pt5; http://dx.doi.org/10.1126/scisignal.2002258; PMID: 21868360
- Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Invest 2010; 120:4141 - 54; http://dx.doi.org/10.1172/JCI42980; PMID: 20972335
- Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, et al. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun 2011; 2:554; http://dx.doi.org/10.1038/ncomms1555; PMID: 22109528
- Noh H, Hong S, Dong Z, Pan ZK, Jing Q, Huang S. Impaired MicroRNA Processing Facilitates Breast Cancer Cell Invasion by Upregulating Urokinase-Type Plasminogen Activator Expression. Genes Cancer 2011; 2:140 - 50; http://dx.doi.org/10.1177/1947601911408888; PMID: 21779487
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 2006; 66:9090 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-06-1403; PMID: 16982751
- Banerjee N, Talcott S, Safe S, Mertens-Talcott SU. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res Treat 2012; 136:21 - 34; http://dx.doi.org/10.1007/s10549-012-2224-0; PMID: 22941571
- Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res 2004; 64:8585 - 94; http://dx.doi.org/10.1158/0008-5472.CAN-04-1136; PMID: 15574765
- Debnath J, Walker SJ, Brugge JS. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J Cell Biol 2003; 163:315 - 26; http://dx.doi.org/10.1083/jcb.200304159; PMID: 14568991
- Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, et al. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci U S A 2004; 101:1257 - 62; http://dx.doi.org/10.1073/pnas.0308090100; PMID: 14739340
- Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 2003; 1:e012; http://dx.doi.org/10.1621/nrs.01012; PMID: 16604184
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286:531 - 7; http://dx.doi.org/10.1126/science.286.5439.531; PMID: 10521349
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19:185 - 93; http://dx.doi.org/10.1093/bioinformatics/19.2.185; PMID: 12538238
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol 2006; 411:134 - 93; http://dx.doi.org/10.1016/S0076-6879(06)11009-5; PMID: 16939790
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001; 98:5116 - 21; http://dx.doi.org/10.1073/pnas.091062498; PMID: 11309499
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15 - 20; http://dx.doi.org/10.1016/j.cell.2004.12.035; PMID: 15652477