Abstract
The latest studies of the interactions between oncogenes and its target cell have shown that certain oncogenes may act as passengers to reprogram tissue-specific stem/progenitor cell into a malignant cancer stem cell state. In this study, we show that the genetic background influences this tumoral stem cell reprogramming capacity of the oncogenes using as a model the Sca1-BCRABLp210 mice, where the type of tumor they develop, chronic myeloid leukemia (CML), is a function of tumoral stem cell reprogramming. Sca1-BCRABLp210 mice containing FVB genetic components were significantly more resistant to CML. However, pure Sca1-BCRABLp210 FVB mice developed thymomas that were not seen in the Sca1-BCRABLp210 mice into the B6 background. Collectively, our results demonstrate for the first time that tumoral stem cell reprogramming fate is subject to polymorphic genetic control.
Introduction
Recent contributions have introduced a new perspective on oncogenic transformation, where certain oncogenes may act as passengers to reprogram tissue-specific stem/progenitor cell into a malignant cancer stem cell state.Citation1,Citation2 The existence of this tumoral stem cell reprogramming as a cancer driver seems to be a common intrinsic mechanism for many types of cancerCitation3-Citation16 and opens a clear hope for cancer treatment, since epigenetic modifications, unlike genetic changes, can be erased, manipulated, and reinitiated. If we can understand the regulation of the oncogene–target cell interaction, and if, as a result, we learn how to manipulate cellular states experimentally, we could unlock the potential to provide great advances in human cancer medicine.Citation1 Thus, one of the most essential cellular substratum that would determine cancer behavior and evolution variability among patients would be the reprogramming potential of the oncogenic event. Therefore, we hypothesized that this tumor reprogramming fate at the cancer stem cell (CSC) level should be the target where, at least partly, the action of those genetic determinants takes place, such as modifier genes, which determine the differences among individuals regarding tumor susceptibility, treatment response, and evolution.
In order to probe whether the genetic background could influence the tumoral stem cell reprogramming fate, we have used as a model the Sca1-BCR-ABLp210 mice, because the type of tumor that develops is a function of stem cell reprogramming.Citation4-Citation8,Citation17 In this model the restricted expression of the BCR-ABL oncogene, linked to chronic myeloid leukemia (CML) disease, to the hematopoietic stem cell compartment is capable of generating a full-blown tumor with all its differentiated cellular components, showing a hands-off role for BCR-ABL in regulating CML formation.Citation4-Citation8,Citation17 BCR-ABL oncogene inactivation could not change this tumor reprogramming fate at the CSC level, in agreement with the common occurrence of tumor relapse by which human CML evolves to escape BCR-ABL pharmacological inactivation.Citation18-Citation21 These observations suggest that the susceptibility to development of CML is intrinsic to the BCR-ABLp210-induced reprogramming of stem cells. As a first step toward identification of genes that control tumor stem cell reprogramming susceptibility, Sca1-BCRABLp210 B6/FVB F1 hybrid mice and pure FVB mice were generated and tested for CML development. The Sca1-BCRABLp210 B6/FVB F1 hybrid mice carry modifier alleles conferring resistance to the development of CML. However, the pure FVB mice carry modifier alleles that result in the rapid development of lethal thymomas. Remarkably, the data provide evidence for the first time that modifier loci would then determine the fate of the oncogene–target cell interaction.
Results and Discussion
In order to probe if the genetic background could affect the tumoral stem cell reprogramming fate, we have taken advantage of our Sca1-BCRABLp210 mouse model of human chronic myeloid leukemia (CML). This transgenic mouse was engineered to express the human BCRABLp210 cDNA under the control of the Sca1 promoter in order to limit and determine the effect of ectopic expression of BCRABLp210 in hematopoietic stem/progenitor cells.Citation4-Citation8,Citation17 This model not only faithfully recapitulates the human disease, but also has been able to anticipate that human CML stem cells’ survival is Bcr-Abl kinase-independent, and suggests that curative approaches in CML must focus on kinase-independent mechanisms of resistance.Citation18-Citation21 Thus, this model represents an ideal system to analyze the contributions of the genetic background to the fate of the interaction of the BCR-ABLp210 oncogene and the target cell, because the type of tumor that develops is a function of tumoral stem cell reprogramming.Citation4-Citation8,Citation17 To this end, the Sca1-BCRABLp210 transgene was moved from the B6 to the FVB genetic background through 6 generations of backcrossing in our laboratory. The resulting strain (FVB, Sca1-BCRABLp210) was used for experiments described in this study. B6/FVB F1 mice used in this study were obtained by breeding the FVB, Sca1-BCRABLp210 mice with regular B6 mice.
In the Sca1-BCRABlp210 model, the type of tumor that develops is a function of stem cell reprogramming. The modifier loci would then determine the fate of this interaction. Mice were monitored clinically and by serial peripheral blood count for evidence of CML for 24 months. As described, all B6, Sca1- BCRABLp210 mice develop CML ().Citation4-Citation8,Citation17 Surprisingly, when the Sca1-BCRABLp210 B6/FVB F1 mice were analyzed, CML was not found in the majority of them, and the survival of these Sca1-BCRABLp210 B6/FVB F1 mice was significantly increased in comparison with B6, Sca1-BCRABLp210 mice (). On the contrary, the majority of old Sca1-BCRABLp210 B6/FVB F1 mice do not develop CML as evidenced by the normal spleen sizes and normal leukocyte cellularity in the peripheral blood. The absence of CML disease was further confirmed by histologic analysis, which revealed normal spleen where we cannot detect the dramatic expansion of progenitors and differentiated myeloid cells that is characteristic of CML. Quantitative RT-PCR of BCR-ABLp210 messenger mRNA confirmed that BCR-ABL was expressed in Sca1-BCRABLp210 B6/FVB F1 hematopoietic stem cells (data not shown). These results indicate that the FVB genetic component in hematopoietic stem cells does interfere with the development of CML induced by BCR-ABL.
Table 1. Genetic background affects stem cell reprogramming in Sca1-BCRABLp210 mice
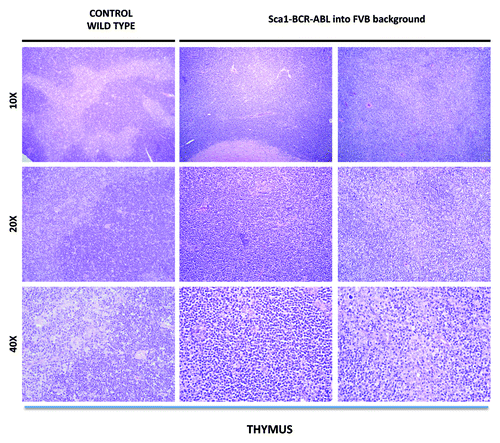
In contrast, Sca1-BCRABLp210 mice inbred into the FVB background are comparatively resistant to the development of CML, as are Sca1-BCRABLp210 B6/FVB(F1) hybrid mice (). However, all FVB, Sca1-BCRABLp210 mice spontaneously developed lymphomas that were not seen in the Sca1-BCRABLp210 mice, either into the hybrid B6/FVB background or into the B6 background. Lymphomas were observed as early as 16 weeks of age and reached an incidence of 100% by 40 weeks of age (). FVB, Sca1-BCRABLp210 mice with lymphomas have typically enlarged thymus comparing to age-matched wild-type controls (). Therefore, genes in the FVB genome can significantly increase the incidence of lymphoma and accelerate the disease when compared with the B6 or FVB/B6 F1 mice. The lymphomas in FVB, Sca1-BCRABLp210 mice were composed of blastic lymphoid cells that effaced the normal architecture of the thymus. The cellular morphology and the pattern of involvement of lymphoid organs mimic the features of lymphoblastic lymphoma in humans ().
Figure 1. Macroscopic appearance of the thymus in Sca1-BCRABLp210 mice in FVB background. Sca1-BCRABLp210 mice in FVB background with signs of disease were analyzed and the thymus was surgically removed. Macroscopically there was sign of thymoma. A thymus of an aged wild-type mouse is shown as a control.
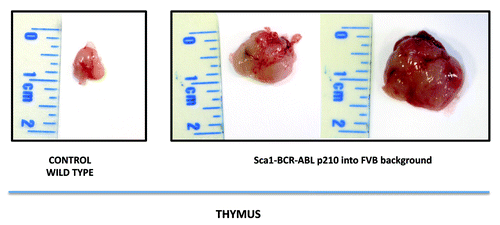
Figure 2. Representative histologic appearance of thymus of diseased Sca1-BCR-ABLp210 into FVB background and control wild-type mice after hematoxylin-eosin staining. Note the organ infiltration by blast lymphoid cells.
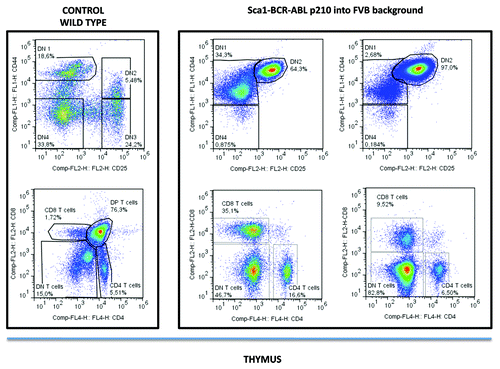
Thymocytes are classified into a number of distinct maturational stages based on the expression of cell surface markers. The earliest thymocyte stage is the double-negative stage (negative for both CD4 and CD8), which can be divided into four substages using CD25 and CD44 markers. The next major stage is the double-positive stage (positive for both CD4 and CD8). The final stage in maturation is the single positive stage (positive for either CD4 or CD8). The analysis of the lymphomas of FVB, Sca1-BCRABLp210 mice by flow cytometry showed predominantly the presence of CD44+CD25+CD4−CD8− double-negative T cells in the thymus (). Thus the genetic background influences the tumoral stem cell reprogramming ability of the BCRABLp210 oncogene.
Figure 3. Phenotypes of lymphomas in Sca1-BCR-ABLp210 into FVB background. All lymphomas analyzed had a similar phenotype with predominantly CD4−CD8− double-negative thymocytes. The use of CD25 and CD44 lineage markers allowed to identify these CD4−CD8− double negative thymocytes as DN2 (CD44+CD25+) thymocytes.
Overall, our work demonstrates for the first time that modifier loci would then determine the fate of the tumoral stem cell reprogramming. These studies represent a first step toward the identification of genes that modify stem cell reprogramming susceptibility in Sca1-BCRABLp210 mice. Because hematopoietic tumors can be quantified and classified relatively easily, the mapping of the modifiers that control stem cell reprogramming should be possible. Molecular identification of such loci should lead to a better understanding of stem cell reprogramming development. These findings have important implications, as they suggest that it would be possible to prevent cancer to develop once a cancer stem cell has been created.
Materials and Methods
Ethics statement
All animal work has been conducted according to relevant national and international guidelines and it has been approved by the Bioethics Committee of University of Salamanca and by the Bioethics Subcommittee of Consejo Superior de Investigaciones Cientificas (CSIC).
Mice
The Sca1-BCRABLp210 transgene was under the control of the Sca1 promoter as previously described.Citation4 The transgene was moved from the B6 to the FVB genetic background through 6 generations of backcrossing in our laboratory. The resulting strain (FVB, Sca1-BCRABLp210) was used for experiments described in this study. B6/FVB F1 mice used in this study were obtained by breeding the FVB, Sca1-BCRABLp210 mice with regular B6 mice from the Jackson Laboratory. Mice were monitored for tumors through physical examination. Mice showing sufficient signs of pain and suffering including thymic and cervical lymphoid enlargement were euthanized for a complete necropsy. Final diagnosis of disease was established by clinical, pathologic, and FACS analysis. Mice without tumors were monitored for 2 y and then were also euthanized for a complete necropsy.
Histological analysis
All mice included in this study were subjected to standard necropsy. All major organs were examined under the dissecting microscope, and samples of each organ were processed into paraffin, sectioned, and examined histologically. All tissue samples were taken by the pathologist from homogenous and viable portions of the resected sample and fixed within 2–5 min. of excision. Hematoxylin and eosin-stained sections of each tissue were reviewed by a single pathologist (OB). For comparative studies, age-matched mice were used.
Real-time PCR quantification
cDNA for use in quantitative PCR studies was synthesized using reverse transcriptase (Access RT-PCR System; Promega). Two µl of second round amplified RNA was transcribed. Primers and probes used for quantitative PCR are commercially available (TaqMan Assays-on-Demand Gene Expression Products, Applied Biosystems). In addition the probes were designed so that genomic DNA would not be detected during the PCR. The sequences of the specific primers and probes were as follow: BCR-ABLp210, sense primer 5′-TTCTGAATGTCATCGTCCACTCA-3′, antisense primer 5′-AGATGCTACTGGCCGCTGA-3′ and probe 5′-CCACTGGATTTAAGCAGAGTTCAAAAGCCC-3′; c-Abl, sense primer 5′-CACTCTCAGCATCACTAAAGGTGAA-3′, antisense primer 5′-CGTTTGGGCTTCACACCATT-3′, and probe 5′-CCGGGTCTTGGGTTATAATCACAATG-3′.
Analysis and monitoring of disease
Peripheral blood was collected from retro-orbital plexus with a heparinized capillary tube, and total white blood cell and differential counts were performed twice a week. The number of white blood cells was determined with a hemocytometer after lysis of enucleated red blood cells with RCLB lysis buffer (0.15 M NH4Cl; 1 mM KHCO3; 0.1 mM Na2-EDTA, pH 7.4).
Flow cytometry
Nucleated cells were obtained from total bone marrow (flushing from the long bones), peripheral blood, thymus, liver, and spleen. In order to prepare cells for flow cytometry, contaminating red blood cells were lysed with RCLB lysis buffer and the remaining cells were then washed in PBS with 2% FCS. After staining, all cells were washed once in PBS with 2% FCS containing 2 mg/mL propidium iodide (PI) to allow dead cells to be excluded from both analyses and sorting procedures. Monoclonal antibodies were obtained from PharMingen and included: lineage markers (CD45R/B220, for B lineage staining; CD4, CD8 and CD44 and CD25 for T cell lineage; CD11b and Gr1 for myeloid lineage and TER119 for erythroid lineage) and Sca1 (E13–161.7) for stem cells. Single-cell suspensions from the different tissue samples obtained by routine techniques were incubated first with purified anti-mouse CD32/CD16 (PharMingen) prior to the addition of other antibodies, to block binding via Fc receptors and then with an appropriate dilution of the different antibodies at room temperature or 4°C, respectively. The samples and the data were analyzed in a FACSCalibur using CellQuest software (Becton Dickinson). Specific fluorescence of FITC and PE excited at 488 nm (0.4 W) and 633 nm (30 mW), respectively, and known forward and orthogonal light-scattering properties of mouse cells were used to establish gates. For each analysis a total of at least 5000 viable (PI−) cells were assessed.
Acknowledgments
We are indebted to all members of lab 13 and lab 7 at IBMCC for useful discussions and for his critical reading of the manuscript. Research in ISG group is partially supported by FEDER and by MICINN (SAF2012–32810), by MEC OncoBIO Consolider-Ingenio 2010 (Ref. CSD2007–0017), by NIH grant (R01 CA109335–04A1), the ARIMMORA project (FP7-ENV-2011, European Union Seventh Framework Program) and by proyecto en red de investigación en celulas madre tumorales supported by Obra Social Kutxa y Conserjería de Sanidad de la Junta de Castilla y Leon. All Spanish funding is co-sponsored by the European Union FEDER program. ISG lab is a collaborator of the EuroSyStem and the DECIDE Network funded by the European Union under the FP7 program.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Vicente-Dueñas C, Romero-Camarero I, Cobaleda C, Sánchez-García I. Function of oncogenes in cancer development: a changing paradigm. EMBO J 2013; 32:1502 - 13; http://dx.doi.org/10.1038/emboj.2013.97; PMID: 23632857
- Cobaleda C, Sánchez-García I. Back to the beginning: The initiation of cancer. Bioessays 2013; 35:413; http://dx.doi.org/10.1002/bies.201300024; PMID: 23553938
- Sánchez-García I, Vicente-Dueñas C, Cobaleda C. The theoretical basis of cancer-stem-cell-based therapeutics of cancer: can it be put into practice?. Bioessays 2007; 29:1269 - 80; http://dx.doi.org/10.1002/bies.20679; PMID: 18022789
- Pérez-Caro M, Cobaleda C, González-Herrero I, Vicente-Dueñas C, Bermejo-Rodríguez C, Sánchez-Beato M, et al. Cancer induction by restriction of oncogene expression to the stem cell compartment. EMBO J 2009; 28:8 - 20; http://dx.doi.org/10.1038/emboj.2008.253; PMID: 19037256
- Vicente-Dueñas C, Pérez-Caro M, Abollo-Jiménez F, Cobaleda C, Sánchez-García I. Stem-cell driven cancer: “hands-off” regulation of cancer development. Cell Cycle 2009; 8:1314 - 8; http://dx.doi.org/10.4161/cc.8.9.8217; PMID: 19279406
- Sánchez-García I. The crossroads of oncogenesis and metastasis. N Engl J Med 2009; 360:297 - 9; http://dx.doi.org/10.1056/NEJMcibr0808031; PMID: 19144947
- Sánchez-García I. Getting to the stem of cancer. Semin Cancer Biol 2010; 20:63 - 4; http://dx.doi.org/10.1016/j.semcancer.2010.05.001; PMID: 20451615
- Castellanos A, Vicente-Dueñas C, Campos-Sánchez E, Cruz JJ, García-Criado FJ, García-Cenador MB, et al. Cancer as a reprogramming-like disease: implications in tumor development and treatment. Semin Cancer Biol 2010; 20:93 - 7; http://dx.doi.org/10.1016/j.semcancer.2010.02.001; PMID: 20188174
- González-Herrero I, Vicente-Dueñas C, Orfao A, Flores T, Jiménez R, Cobaleda C, et al. Bcl2 is not required for the development and maintenance of leukemia stem cells in mice. Carcinogenesis 2010; 31:1292 - 7; http://dx.doi.org/10.1093/carcin/bgq062; PMID: 20299524
- Vicente-Dueñas C, Barajas-Diego M, Romero-Camarero I, González-Herrero I, Flores T, Sánchez-García I. Essential role for telomerase in chronic myeloid leukemia induced by BCR-ABL in mice. Oncotarget 2012; 3:261 - 6; PMID: 22408137
- Vicente-Dueñas C, Fontán L, Gonzalez-Herrero I, Romero-Camarero I, Segura V, Aznar MA, et al. Expression of MALT1 oncogene in hematopoietic stem/progenitor cells recapitulates the pathogenesis of human lymphoma in mice. Proc Natl Acad Sci U S A 2012; 109:10534 - 9; http://dx.doi.org/10.1073/pnas.1204127109; PMID: 22689981
- Vicente-Dueñas C, Cobaleda C, Martínez-Climent JÁ, Sánchez-García I. MALT lymphoma meets stem cells. Cell Cycle 2012; 11:2961 - 2; http://dx.doi.org/10.4161/cc.21264; PMID: 22825253
- Vicente-Dueñas C, Romero-Camarero I, González-Herrero I, Alonso-Escudero E, Abollo-Jiménez F, Jiang X, et al. A novel molecular mechanism involved in multiple myeloma development revealed by targeting MafB to haematopoietic progenitors. EMBO J 2012; 31:3704 - 17; http://dx.doi.org/10.1038/emboj.2012.227; PMID: 22903061
- Vicente-Dueñas C, Romero-Camarero I, García-Criado FJ, Cobaleda C, Sánchez-García I. The cellular architecture of multiple myeloma. Cell Cycle 2012; 11:3715 - 7; http://dx.doi.org/10.4161/cc.22178; PMID: 22983005
- Vicente-Dueñas C, González-Herrero I, García Cenador MB, García Criado FJ, Sánchez-García I. Loss of p53 exacerbates multiple myeloma phenotype by facilitating the reprogramming of hematopoietic stem/progenitor cells to malignant plasma cells by MafB. Cell Cycle 2012; 11:1479 - 80; http://dx.doi.org/10.4161/cc.22186; PMID: 22487683
- Romero-Camarero I, Jiang X, Natkunam Y, Lu X, Vicente-Dueñas C, Gonzalez-Herrero I, et al. Germinal centre protein HGAL promotes lymphoid hyperplasia and amyloidosis via BCR-mediated Syk activation. Nat Commun 2013; 4:1338; http://dx.doi.org/10.1038/ncomms2334; PMID: 23299888
- Velasco-Hernández T, Vicente-Dueñas C, Sánchez-García I, Martin-Zanca D. p53 restoration kills primitive leukemia cells in vivo and increases survival of leukemic mice. Cell Cycle 2013; 12:122 - 32; http://dx.doi.org/10.4161/cc.23031; PMID: 23255106
- Chomel JC, Bonnet ML, Sorel N, Bertrand A, Meunier MC, Fichelson S, et al. Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood 2011; 118:3657 - 60; http://dx.doi.org/10.1182/blood-2011-02-335497; PMID: 21791426
- Chu S, McDonald T, Lin A, Chakraborty S, Huang Q, Snyder DS, et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood 2011; 118:5565 - 72; http://dx.doi.org/10.1182/blood-2010-12-327437; PMID: 21931114
- Kumari A, Brendel C, Hochhaus A, Neubauer A, Burchert A. Low BCR-ABL expression levels in hematopoietic precursor cells enable persistence of chronic myeloid leukemia under imatinib. Blood 2012; 119:530 - 9; http://dx.doi.org/10.1182/blood-2010-08-303495; PMID: 22101898
- Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood 2012; 119:1501 - 10; http://dx.doi.org/10.1182/blood-2010-12-326843; PMID: 22184410