Abstract
Maintaining genome stability is crucial for all cells. The budding yeast Elg1 protein, the major subunit of a replication factor C-like complex, is important for genome stability, since cells lacking Elg1 exhibit increased recombination and chromosomal rearrangements. This genome maintenance function of Elg1 seems to be conserved in higher eukaryotes, since removal of the human Elg1 homolog, encoded by the ATAD5 gene, also causes genome instability leading to tumorigenesis. The fundamental molecular function of the Elg1/ATAD5-replication factor C-like complex (RLC) was, until recently, elusive, although Elg1/ATAD5-RLC was known to interact with the replication sliding clamp PCNA. Two papers have now reported that following DNA replication, the Elg1/ATAD5-RLC is required to remove PCNA from chromatin in yeast and human cells. In this Review, we summarize the evidence that Elg1/ATAD5-RLC acts as a PCNA unloader and discuss the still enigmatic relationship between the function of Elg1/ATAD5-RLC in PCNA unloading and the role of Elg1/ATAD5 in maintaining genomic stability.
Keywords: :
The Elg1/ATAD5 Replication Factor C-like Complex: A Mysterious Guardian of Genomic Stability
The ELG1 gene (enhanced level of genomic instability 1) was first identified in Saccharomyces cerevisiae as required for accurate chromosome maintenance, a role found to be shared by its human homolog, called ATAD5 (ATPase family AAA domain-containing protein 5). Budding yeast cells lacking Elg1 exhibit pleiotropic chromosome instability phenotypes including increased recombination rate, gross chromosomal rearrangements, elongated telomeres, cohesion defects, and sensitivity to the DNA-alkylating drug MMS.Citation1-Citation9 The role of Elg1 in maintaining chromosome stability seems to be conserved in higher eukaryotes, since mutating the ATAD5 gene caused genomic instability and tumorigenesis in mice and human cells.Citation10,Citation11 Elg1 and ATAD5 were identified as the major subunits of a replication factor C-like complex (RLC) having structural similarity to replication factor C (RFC), which loads the polymerase clamp PCNA at replication forks. The Elg1/ATAD5-RLC physically interacts with PCNA, but its physiological role in PCNA transactions for years remained obscure, making the Elg1-RLC a particularly mysterious guardian of genomic stability. Two recent papers have illuminated the in vivo effect of Elg1/ATAD5 on PCNA. Anne Donaldson’s group reported that the Elg1-RLC functions in PCNA unloading from chromatin during replication in yeast,Citation12 while Kyungjae Myung’s groups showed that ATAD5 is needed for proper removal of PCNA and disassembly of replication factories in human cell lines.Citation13 In this review, we focus on these recent advances and discuss how loss of Elg1 function might cause genome instability. We begin by briefly introducing PCNA and previous investigations of the effects of RLCs. We then summarize advances in the 2 recent papers and outline possible PCNA unloading mechanisms. Finally, we discuss how failure of PCNA unloading might impact on genomic stability.
Role of PCNA in DNA Replication
A central coordinator of DNA replication, PCNA is a homotrimeric, ring-shaped molecule that encircles DNA to act as a polymerase clamp as well as a sliding platform for recruitment of other replication and repair proteins, including DNA helicase, nuclease, ligase, and histone chaperones.Citation14 On the lagging strand, PCNA cooperates with DNA polymerase δ to synthesize DNA discontinuously in a series of Okazaki fragments, 100–200 nucleotides in length. Ligation of the Okazaki fragments into a continuous daughter strand then depends on PCNA-mediated recruitment of the flap endonuclease FEN-1 and DNA ligase I.Citation15
On the lagging strand, PCNA must be loaded repeatedly onto the DNA to synthesize each Okazaki fragment. PCNA is loaded by RFC, a hetero-pentameric complex consisting of Rfc1–5 subunits.Citation16-Citation18 The five subunits show sequence similarity to each other and are members of the AAA+ ATPase family. On binding ATP, RFC interacts with PCNA and opens its trimeric ring. The open PCNA-RFC complex then recognizes and loads at the 3′ end of a primer–template junction. On hydrolysis of ATP, RFC ejects PCNA, leaving it on the DNA in closed form. The loading process is discussed in detail in excellent recent reviews.Citation19,Citation20
RFC-like complexes
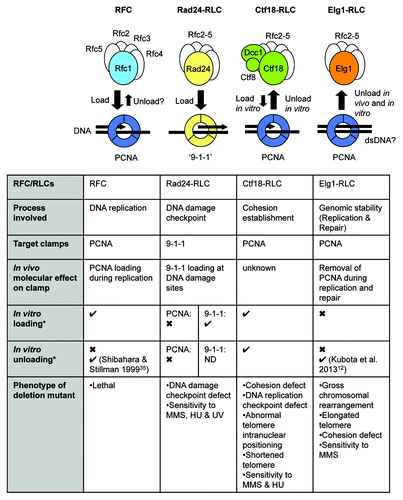
All eukaryotic cells contain a series of 3 RFC-like complexes (RLCs), which share with RFC the Rfc2–5 subunits, but in place of Rfc1 have an alternative subunit: Rad24 (called Rad17 in human), Ctf18, or Elg1 (ATAD5 in human) ().Citation21 The structural resemblance of these RLCs with RFC suggested they may mediate interactions of ring-shaped clamps with DNA—and, indeed, Rad24-RLC, the best-understood of the three RLCs, acts to load the trimeric PCNA-like “9-1-1” complex at damaged DNA for checkpoint activation.Citation22-Citation25 The 9-1-1 complex is conserved, with subunits called RAD9-HUS1-RAD1 in human and Ddc1-Mec3-Rad17 in S. cerevisiae. Ctf18-RLC is unique in the RLC family in forming a heptamer containing 2 additional subunits, Dcc1 and Ctf8.Citation26,Citation27 Ctf18-RLC is important for sister chromatid cohesion and replication checkpoint activationCitation26,Citation28,Citation29 and is also required for proper telomere length regulation and telomere intranuclear positioning.Citation30 Using in vitro assays in biochemically defined systems, Ctf18-RLC was shown to be capable of loading PCNA onto DNA and also of unloading it,Citation31 but whether the Ctf18-RLC loads or unloads PCNA in vivo remains unclear.
Figure 1. Summary of known functions of replication factor C and replication factor C-like complexes. *Except where indicated, in vitro loading and unloading results as derived from Bylund and Burgers 2005Citation31; Majka and Burgers 2003.Citation24 ND, not determined.
The Elg1/ATAD5-RLC was for years the most mysterious of the 3 RLCs. Although shown to physically interact with PCNA, in vitro biochemical assays suggested that Elg1-RLC could not load PCNA onto or unload it from DNA,Citation6,Citation31 so its fundamental molecular function remained elusive.
Investigation of PCNA Unloading by RFC and RLCs In Vitro
PCNA must be loaded for synthesis of each Okazaki fragment and unloaded on its completion, as well as at replication fork termination. It was proposed that RFC acts to unload PCNA as well as loading it; here, we summarize the in vitro studies that led to this suggestion. In 1996, the Hurwitz and O’Donnell groups reported that human RFC unloads PCNA from singly nicked circular plasmid, as tested using an in vitro system which uses gel filtration to measure free PCNA released from a PCNA-plasmid DNA complex.Citation32,Citation33 However, in a later investigation of PCNA ring opening by yeast RFC, the O’Donnell group made the surprising observation that a tetrameric Rfc2-Rfc5 subassembly (lacking Rfc1) is also capable of opening and unloading PCNA from nicked circular plasmid.Citation34 Since all RLCs contain Rfc2–5, this finding might suggest that all the RLCs as well as RFC could potentially open and unload PCNA, although such a lack of specificity seems unlikely in vivo. PCNA unloading by Elg1-RLC was not tested in this system.
Shibahara and Stillman also showed that human RFC can mediate unloading of PCNA.Citation35 Using cell extract and a plasmid containing the SV40 origin, they found that in the presence of RFC, PCNA is removed from fully replicated SV40 plasmid DNA in an ATP-dependent manner.Citation35 The regulation of PCNA transactions in this SV40 replication system might, however, differ from those in normal genome replication.
The above investigations suggest that RFC can unload PCNA from DNA. However, using an assay similar to that used by the O’Donnell group, Peter Burgers’ group could not detect significant unloading of PCNA by yeast RFC, even on testing several DNA substrates with various concentrations of proteins.Citation31 The Rfc2–5 subcomplex was also unable to unload PCNA in their assay. Their failure to measure significant unloading suggests that, at least in the case of yeast, the efficiency of RFC as a PCNA loader far outweighs its efficiency as an unloader. While it is possible that RFC catalyzes cycles of PCNA loading and unloading, the current evidence does leave significant doubt about whether the canonical RFC complex is a major PCNA unloader in vivo.
In the same in vitro investigation, Bylund and Burgers found that neither Rad24-RLC nor Elg1-RLC were able to mediate significant unloading of PCNA.Citation31 Remarkably, they did find that yeast Ctf18-RLC can unload PCNA from DNA in a reaction driven by ATP hydrolysis.Citation31 However, Ctf18-RLC is probably not the main PCNA unloader in vivo, since ctf18Δ cells or human cells subject to siRNA knockdown of CTF18 expression show approximately normal levels of PCNA on chromatin in an unperturbed S phase.Citation13,Citation29 Its effectiveness in unloading PCNA from DNA in vitroCitation31 does hint that Ctf18-RLC may play this role under specific in vivo circumstances yet to be elucidated.
These results are summaried in . Taken together, previous studies certainly do not exclude that RFC might unload PCNA during DNA replication, but they have equally failed to demonstrate that RFC acts as the major PCNA unloader in the normal in vivo situation. The need for RFC to load PCNA has made it difficult to design experiments that might test for unloading of PCNA by RFC in vivo.
PCNA Unloading by S. cerevisiae Elg1-RLC
Several lines of evidence from yeast suggested a role for Elg1-RLC in PCNA regulation during DNA replication.Citation6,Citation29,Citation36 Martin Kupiec’s group demonstrated that yeast Elg1-RLC preferentially interact with SUMOylated PCNA, and in cells lacking Elg1, they found SUMOylated PCNA accumulated on chromatin in the presence of the DNA-alkylating drug MMS,Citation36 suggesting that Elg1-RLC might unload SUMOylated PCNA from DNA. Using quantitative proteomics, Anne Donaldson’s group showed that in yeast cells lacking Elg1, PCNA accumulates abnormally on chromatin in mid-S phase even without MMS treatment.Citation29,Citation37 These studies suggested PCNA unloading as a possible role for Elg1-RLC in vivo, but could not exclude the possibility that PCNA accumulation on chromatin might be an indirect effect of the elg1Δ mutation.
The new paper from the Donaldson group provides more direct evidence that yeast Elg1-RLC functions in PCNA unloading during DNA replication ().Citation12 This study used an improved degron system to test the immediate effects of Elg1 depletion, revealing that PCNA accumulates on chromatin during the first S phase without Elg1. This accumulated PCNA is unloaded rapidly in vivo if Elg1 expression is switched back on, either during or after completion of DNA replication. Overexpression of Rfc1, Ctf18, or Rad24 could not substitute for Elg1 in removing accumulated PCNA. These results implicated Elg1 in PCNA unloading in vivo. The investigators then proceeded to demonstrate PCNA unloading by Elg1-RLC using a novel in vitro assay, in which chromatin prepared from an elg1Δ mutant was treated with affinity-purified Elg1-RLC, resulting in removal of PCNA. While formal identification of Elg1-RLC as a PCNA unloader will require reconstitution of the reaction with biochemically pure ingredients, this study provides the first coupled in vivo and in vitro evidence for PCNA removal by a clamp unloader.
The complex nature of the chromatin substrate used by Kubota et al.Citation12 leaves important questions about the biochemistry of PCNA unloading by Elg1-RLC. The fact that Elg1-RLC could not unload PCNA in the earlier biochemical assay using defined componentsCitation31 suggests that there are additional requirements still to be identified that are needed for Elg1-RLC to unload PCNA from DNA. The earlier assay used purified PCNA loaded at a synthetic 3′ primer–template junction,Citation31 while use of native chromatin from elg1Δ as substrate may provide PCNA in a form that is more suitable for unloading. In particular, in the Donaldson group’s system, the PCNA may be modified, histones can be deposited onto DNA, and any co-factors supporting Elg1-dependent PCNA unloading are likely to be present on chromatin. Alternatively, DNA structure might be critical for PCNA unloading by Elg1-RLC. For example, Elg1-RLC might be unable to unload PCNA from a nicked DNA substrate and may remove PCNA only from fully double-stranded DNA, as present after Okazaki fragment processing. It will be of interest to investigate the specific components or requirements needed for PCNA unloading by Elg1-RLC.
PCNA Removal by Human Elg1/ATAD5-RLC
Remarkably, an independent, simultaneous study provided compelling evidence that the function of Elg1/ATAD5 in PCNA unloading is conserved in human cells. This investigation from Kyungjae Myung’s group demonstrated that human ATAD5 is critical for correct regulation of the lifespan of DNA replication factories.Citation13 Specifically, knocking down ATAD5 in human cell lines caused PCNA to accumulate on chromatin and remain there even after replication at that site had finished—analogous to the retention of PCNA on chromatin seen by Kubota et al.Citation12 in a yeast elg1Δ mutant and consistent with the idea that Elg1/ATAD5-RLC unloads PCNA from DNA. Prolonged PCNA retention on chromatin caused by the knockdown of ATAD5 resulted in an extended lifespan of DNA replication factories, with all replication proteins enriched in the long-lived factories. This effect correlated with an accumulation on chromatin of other replisome proteins, including MRN complex and the chromatin-remodeling protein, CAF1. This accumulation was mediated by the excess PCNA retention, since simultaneous ATAD5 and PCNA knockdown mitigated the retention of other replisome components. As well as half-life, the intensity and number of DNA replication factories were increased upon knockdown of ATAD5. Interestingly, in the absence of ATAD5, many DNA replication factories were inactive for DNA synthesis, remaining even during G2 phase of cell cycle. Conversely, ectopic ATAD5 expression caused a reduction of PCNA on chromatin, consistent with the idea that ATAD5 drives PCNA unloading. Although PCNA unloading by human ATAD5-RLC has yet to be directly demonstrated in vitro, the 2 recent studies by Donaldson’s and Myung’s groups together provide good evidence that the Elg1/ATAD5-RLC acts to unload PCNA during DNA replication, this function being conserved from yeast to human.
Does the Elg1/ATAD5-RLC Preferentially Unload SUMOylated PCNA?
Studies from Kupiec’s group suggest the interesting idea that yeast Elg1-RLC is an unloader specific to SUMOylated PCNA. This idea is based on their observations that Elg1-RLC preferentially interacts with SUMOylated PCNA, and that SUMOylated PCNA accumulates on chromatin in elg1Δ mutant, especially in the presence of MMS.Citation36 Kubota et al. also observed an increase of SUMOylated PCNA on chromatin in elg1Δ mutant during unperturbed S phase.Citation12 However, an unSUMOylatable PCNA Pol30-K164R/K127R still accumulates on chromatin when Elg1 is deleted,Citation12 implying that PCNA SUMOylation is not essential for PCNA to be unloaded by Elg1-RLC. Although SUMOylation is not absolutely required for PCNA unloading by Elg1-RLC, the observations by Kubota et al.Citation12 are still consistent with the idea that Elg1-RLC has a preference for unloading SUMOylated PCNA. DNA association leads to PCNA SUMOylation,Citation38 suggesting that the increase of SUMOylated PCNA in an elg1Δ mutant is probably a direct consequence of the prolonged retention of PCNA on chromatin when its normal unloader is absent.
SUMOylation of PCNA has been observed in higher eukaryotes, such as chicken DT40 cells, X. laevis egg extracts, and recently mammalian cells.Citation39-Citation43 In human cell lines, the modification is present at very low levels.Citation43 Human ATAD5-RLC could potentially prefer to interact with SUMOylated PCNA, since ATAD5 has a SUMO interaction motif in its N-terminal domain.Citation44,Citation45 However, accumulation of SUMOylated PCNA on chromatin was not observed in ATAD5-knockdown cells, even though both unmodified and ubiquitinated PCNA increased on chromatin (the latter due to defects in recruiting USP1-UAF1 deubiquitinating enzymes to PCNA).Citation13,Citation44 Overall, it seems likely that yeast Elg1-RLC has a preference, but not an absolute requirement, to unload SUMOylated PCNA, but any such preference appears not to be conserved in human ATAD5-RLC.
Structural Domain Requirements for PCNA Unloading
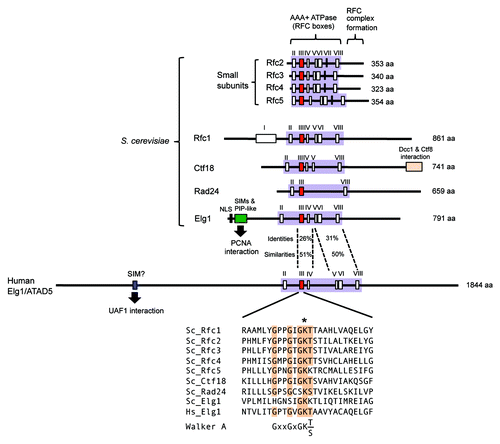
What are the structural features of Elg1/ATAD5 that might permit it to act as a PCNA unloader? As illustrated in , Elg1 has a central domain showing similarity to Rfc1 and Ctf18 and containing a conserved ATPase domain, flanked by unique N-terminal and C-terminal regions. Below we discuss our understanding of the Elg1/ATAD5 structure in relation to its proposed role in PCNA unloading.
Figure 2. Structure of RFC/RLCs. Within the AAA+ ATPase domain (colored lilac), box III (colored red) contains Walker A sequence, box V contains Walker B sequence, box VII (in Rfc2–5 subunits) contains Arginine finger. Detailed sequence comparison of Walker A motifs shown below. *Indicates conserved lysine. NLS, nuclear localization signal; SIM, SUMO interaction motif; PIP-like, PCNA interacting peptide-like.
ATPase domain
All subunits of RFC/RLC subunits (excluding Dcc1 and Ctf8) are members of the AAA+ ATPase family and contain ATP-binding Walker A and B motifs (). Walker A motifs contain an invariant lysine that is critical for ATP binding.Citation46 In human Rfc1, and in yeast Rad24 and Ctf18, this invariant lysine is critical for function,Citation31,Citation47-Citation50 but surprisingly, the lysine in yeast Rfc1 is not essential for PCNA loading in vivo or in vitro.Citation51,Citation52
Human Elg1/ATAD5 also contains this invariant lysine within a consensus Walker A motif (). Myung’s group has tested if the ATPase domain in human Elg1/ATAD5 is important for its function by mutating the invariant lysine (K1138). An ATAD5 K1138E mutant protein could not prevent PCNA retention on chromatin or the defective replication factories of ATAD5 knockdown cells, suggesting the invariant lysine is important for removal of PCNA from chromatin.Citation13 However, since the K1138E mutation also affects the stability of ATAD5 protein and its interaction with small Rfc subunits,Citation13 failure of PCNA removal may not be solely due to loss of ATP binding.
Yeast Elg1 lacks some of the consensus residues of the Walker A motif (). The invariant lysine is present in yeast Elg1 but does not appear to be required for its in vivo function, in contrast with human ATAD5. Davidson and Brown mutated the invariant lysine (Elg1 K343) and found the cells exhibited wild-type levels of sensitivity to MMS (unlike the elg1Δ mutant which shows greatly elevated MMS sensitivity).Citation53 Their study suggests that, similar to yeast Rfc1, ATP binding to yeast Elg1 is not essential for its in vivo function. One possibility is that ATP binding to the smaller subunits Rfc2–5 is sufficient for PCNA loading by RFC and unloading by Elg1-RLC in budding yeast. Overall, current results suggest the requirement for ATP binding to Elg1/ATAD5-RLC may differ among species. It will be of interest to test directly whether ATP binding by yeast Elg1 is required for PCNA unloading.
N-terminal domain
The unique N-terminal domain of Elg1 may make a crucial contribution to PCNA unloading, as it interacts with PCNA and SUMOylated PCNA and is known to be important for in vivo function of Elg1.Citation36,Citation53 Budding yeast Elg1 has 3 SUMO interaction motifs (SIMs) and a PCNA interacting peptide-like (PIP-like) motif in its N-terminal region, conferring a preferential interaction with SUMOylated PCNA ().Citation36 Cells expressing Elg1 lacking the N-terminal 215 amino acids exhibit greater MMS sensitivity than wild type, but less than an elg1Δ mutant.Citation53 These results suggest that the N-terminal region in Elg1 is important for its function in PCNA regulation, and probably in PCNA unloading. Consistent with this idea, yeast cells expressing Elg1 mutated in both SIMs and PIP-like motif also show substantial MMS sensitivity.Citation36
One possibility is that 2 binding modes occur during Elg1-PCNA interaction. First, Elg1-RLC might search for chromatin-bound PCNA/SUMO-PCNA using its N-terminal region, which is probably extended from core “RFC” domain and could act as flexible “scanner”. After docking with its target (i.e., PCNA to be unloaded), Elg1-RLC may then bind PCNA strongly through all 5 subunits (in a manner similar to RFC), and then open the PCNA ring and release it from DNA.
Human ATAD5 has a much longer extended N-terminal region than yeast Elg1, and is therefore larger than yeast Elg1 (1844 a.a. and 791 a.a., respectively) (). Human ATAD5 has a SUMO interaction motif in its N-terminal region, shown to interact with a SUMO-like domain in the deubiquitination factor UAF1.Citation44,Citation45 Human ATAD5 regulates PCNA deubiquitination by recruiting the USP1-UAF1 complex to ubiquitinated PCNA.Citation44 The N-terminal region in human ATAD5 therefore likely confers additional functions in regulation of ubiquitination compared with yeast Elg1. Any PCNA-binding region in the ATAD5 N-terminus has yet to be investigated, and human ATAD5 may also interact with PCNA through its N-terminal region, like yeast Elg1.
C-terminal domain
There is less information about the function of C-terminal domain of yeast Elg1. Loss of the C-terminal 60 amino acids does not affect sensitivity to MMS, except in the context of an N-terminal truncation, where it does somewhat exacerbate MMS sensitivity.Citation53 The C-terminal domain of yeast Elg1 may also therefore contribute to its function, either in PCNA unloading or other cellular roles as discussed below. Interestingly, a C-terminal truncation mutation in ATAD5 found in endometrial tumor caused instability of protein,Citation10 suggesting the ATAD5 C-terminal domain is important to suppress tumorigenesis.
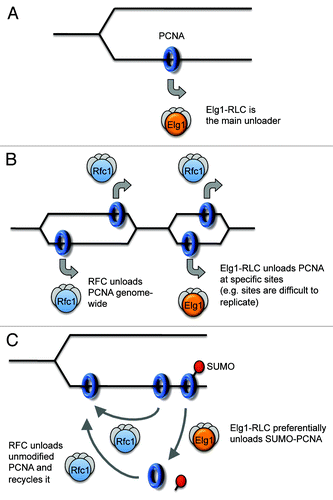
Is Elg1-RLC the Main PCNA Unloader?
The observations described above suggest strongly that Elg1/ATAD5-RLC acts to unload PCNA, but do not address whether Elg1/ATAD5-RLC is the primary PCNA unloader during normal DNA replication. For example, Elg1/ATAD5-RLC might unload PCNA only at specific genomic sites. Three distinct models () are consistent with the published data: (A) Elg1-RLC is the main PCNA unloader. If this is the case then other mechanisms must exist that can eventually unload PCNA, since most PCNA is eventually removed from chromatin after replication even in the absence of Elg1/ATAD5. Without Elg1/ATAD5, PCNA-unloading function could conceivably be taken over by RFC and/or Ctf18-RLC, but with less efficient removal by these complexes resulting in the observed PCNA accumulation on chromatin. It is also possible that PCNA might eventually dissociate from DNA spontaneously without any active unloader.Citation34 (B) RFC could be the major PCNA unloader, with Elg1/ATAD5-RLC as a minor unloader acting at particular sites, for example, sites of cohesion binding or specific sequences that are difficult to replicate. In this case, the accumulation of PCNA on chromatin observed in the absence of Elg1/ATAD5 presumably represents PCNA retained at these particular locations. (C) A further possibility is that RFC is the main unloader of unmodified PCNA, with Elg1-RLC acting primarily to unload SUMOylated PCNA (even though it can also remove unmodified PCNA). In this case, RFC might unload unmodified PCNA from a newly ligated Okazaki fragment and immediately re-load it on a new primer-template junction, allowing rapid recycling of unmodified PCNA. Elg1-RLC might, in contrast, preferentially unload PCNA that has become SUMOylated but not re-load it (due to lack of loading activity). In this case, the PCNA could only be re-loaded after de-SUMOylation and subsequent recognition by RFC, providing a mechanism to prevent loading of SUMOylated PCNA. In fact Models (A) and (C) are not mutually exclusive, since PCNA may usually be SUMOylated by the time it is unloaded, and therefore unloaded by Elg1-RLC in most cases. Current results are consistent with all three possibilities, although the marked PCNA accumulation on chromatin when Elg1/ATAD5 are lacking might be construed as less compatible with its retention only at specific sites (Model B). It will be of interest to test these possibilities directly.
Is PCNA Unloading the Main Cellular Function of the Elg1/ATAD5-RLC?
Budding yeast lacking Elg1 shows pleiotropic phenotypes, including increased chromosomal rearrangements, cohesion defects, elongated telomeres, and sensitivity to MMS ().Citation1-Citation9 Similarly, low expression of ATAD5 in human or mouse cells caused increased chromosomal rearrangements, increased rate of sister chromatid exchange, and mild sensitivity to DNA damaging agents.Citation10,Citation11 In addition, homozygous null mutation of Atad5 in mice caused embryonic lethality (Myung et al., unpublished results). Do all these phenotypes result from compromised PCNA unloading, or do they instead indicate that Elg1/ATAD5 has additional molecular roles beyond PCNA unloading? At this point the answer remains unclear, but the former is a possibility, since, as explained below, these elg1Δ phenotypes could conceivably all result from problems with PCNA transactions. ATAD5 has also been shown to regulate the removal of ubiquitin from PCNA: we also consider possible links between PCNA unloading by ATAD5-RLC, PCNA deubiquitination, and the genome instability observed in mammalian cells in the absence of ATAD5.
Increased chromosomal rearrangements
Could compromised PCNA unloading be an indirect cause of chromosome rearrangements? At least 2 pathways can be envisaged. First, failure of timely PCNA unloading may delay the recycling of PCNA and PCNA-binding proteins, in turn delaying lagging-strand synthesis and resulting in unprocessed lagging-strand replication intermediates that could stimulate inappropriate recombination events, leading ultimately to chromosome rearrangements. Consistent with this idea, cells lacking FEN-1, the flap endonuclease that processes Okazaki fragments, show increased recombination rates and gross chromosomal rearrangements.Citation54,Citation55
A second possible pathway could involve the accumulation on chromatin of PCNA-binding proteins that promote unwanted recombination. One candidate is the AAA+ ATPase Mgs1 that interacts with PCNA. Mgs1 has both PCNA-binding and UBZ (ubiquitin-binding) domains and preferentially interacts with polyubiquitinated PCNA,Citation56 and in an elg1Δ mutant, both ubiquitinated PCNA and Mgs1 are abnormally increased on chromatin.Citation12,Citation29 Polyubiquitinated PCNA is thought to direct error-free repair through template switching,Citation57,Citation58 and while the molecular function of Mgs1 is mysterious, either deletion or overexpression of the MGS1 gene affects recombination and mutation rates.Citation59-Citation61 It therefore seems possible that the PCNA retention on chromatin in elg1Δ leads to an imbalance of Mgs1 recruitment, eventually leading to chromosome rearrangements. In general, it will be of interest to investigate which of the many pathways coordinated by PCNA are affected by loss of Elg1-RLC activity.
Cohesion defect
Cohesion establishment is coupled with DNA replication, and cohesion defects are reported in S. cerevisiae lacking replication proteins, including Ctf4, Ctf18, Mrc1, Tof1, Csm1, and FEN-1.Citation28,Citation62,Citation63 Properly coordinated leading- and lagging-strand progression may be important for cohesion establishment, so that problems with lagging-strand synthesis in elg1Δ cells due to compromised PCNA unloading or recycling might result in cohesion defects. Robert Skibbens’s group has proposed that cohesion establishment occurs in concert with lagging-strand synthesisCitation64—although Frank Uhlmann’s group found no role of the lagging-strand processing enzymes FEN-1 and Dna2 in cohesion establishment.Citation65 A variant of this idea proposes that inefficient PCNA unloading causes progression problems when replication forks encounter cohesin rings, possibly affecting cohesion establishment.
Yet another possibility is that the accumulation of SUMOylated PCNA on chromatin interferes with function of the cohesion establishment factor Eco1. Eco1 normally promotes cohesion by acetylating the Smc3 cohesin subunit during S phase.Citation66-Citation69 Eco1 is recruited by PCNA, and PCNA SUMOylation appears to counteract Eco1 activity.Citation70 It is therefore possible that the excessive PCNA SUMOylation observed in an elg1Δ mutant (probably as a result of extended DNA association) interferes with the function of Eco1 in cohesion establishment. Notably, a pol30–104 mutant (expressing PCNA A251V) that is over-SUMOylated also shows a cohesion defect.Citation70
Elongated telomeres
Again, the elongated telomere phenotype of elg1Δ cells might be connected to lagging-strand synthesis problems caused by compromised PCNA unloading or recycling. Other mutations (pol1, rfc1, pol32Δ, and rad27Δ), affecting lagging-strand synthesis also cause elongated telomeres.Citation71-Citation73 One idea is that delayed lagging-strand synthesis leaves single-stranded DNA exposed and unable to recruit the telomerase inhibitor complex Rap1-Rif1/Rif2 (which binds dsDNA), but still bound by the telomerase-recruiting complex Cdc13-Stn1-Ten1 (which binds to single-strand DNA), promoting telomere extension. Alternatively, PCNA accumulation itself may cause telomere extension by occupying the telomeric DNA and interfering with Rap1-Rif1/Rif2 recruitment, leading to inappropriate extension by telomerase.
MMS sensitivity
We suspect that the MMS sensitivity of elg1Δ cells may be connected to PCNA retention on chromatin, since we find that overexpression of PCNA also causes over-loading of PCNA on chromatin that is coupled with increased MMS sensitivity (unpublished data). The mechanism could result from an imbalance of PCNA modifications important for DNA repair.
Does loss of other activities of Elg1 cause genomic instability?
While speculative, the mechanisms described above suggest plausible pathways, consistent with established observations, through which compromised PCNA unloading might cause the elg1Δ mutant phenotypes. But of course molecular roles of Elg1 unrelated to PCNA unloading could account for some defects of an elg1Δ mutant. In particular, Elg1 interacts with SUMOylated and SIM-containing proteins through poly-SUMO chains, independent of SUMOylated PCNA.Citation74 Disturbing such interactions could affect recombination rate, cohesion establishment, and telomere extension, because SUMOylation is involved in all these processes.Citation75-Citation79 Clearly, further investigations will be needed to shed light on the issue of which elg1Δ phenotypes result from compromised PCNA unloading and which from other Elg1 molecular functions.
Human Elg1/ATAD5 promotes PCNA deubiquitination by recruiting the USP1-UAF1 complex to ubiquitinated PCNA.Citation44 Can failure to deubiquitinate PCNA explain the increased chromosomal rearrangement in ATAD5-knockdown cells? Cells knocked down for USP1 expression show reduced homologous recombination, and a similar effect is observed upon knockdown of ATAD5.Citation44 Thus, the effect of ATAD5 in facilitating PCNA deubiquitination activity may be important during DNA double-strand break-induced homologous recombination. However, studies of mouse mutants suggest that ATAD5 plays a functional role extending well beyond its effect on the deubiquitination system. A null mutation of atad5 in mice causes embryonic lethality (Myung lab, unpublished result), but usp1-null mice are in contrast born (although with an anemic phenotype). No tumorigenesis was observed in the usp1-null mice—in stark contrast to the high incidence of tumorigenesis in atad5 heterozygous mice.Citation10,Citation80 These observations suggest that ATAD5 PCNA unloading activity is important for embryonic development and tumor suppression, in addition to its role in PCNA deubiquitination.
Concluding Remarks and Future Directions
Unloading of PCNA must take place repeatedly on completion of every Okazaki fragment, and also at replication fork termination. Two recent papers have reported that Elg1/ATAD5-RLC functions in removing PCNA from chromatin in yeast and human. These findings represent a substantial advance in understanding the operation of replication forks as well as Elg1/ATAD5 function. Compromised PCNA unloading could impact on multiple chromosome maintenance mechanisms to cause the genome instability of elg1Δ cells, through PCNA accumulation on chromatin, delayed recycling of PCNA, or altered recruitment of PCNA-interacting factors, including cohesion factors and DNA repair proteins.Citation14,Citation29,Citation56 It will be of interest to investigate which PCNA-coordinated pathways are affected by loss of the Elg1-RLC, and the phenotypic consequences, especially in relation to the other activities of Elg1.Citation74
| Abbreviations: | ||
| RLC | = | replication factor C-like complex |
| RFC | = | replication factor C |
Acknowledgments
This work was supported by Cancer Research UK grant A11646 and BBSRC grant BB/K006304/1 to ADD and the intramural research program of the National Human Genome Research Institute to KM.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Maradeo ME, Skibbens RV. The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS One 2009; 4:e4707; http://dx.doi.org/10.1371/journal.pone.0004707; PMID: 19262753
- Parnas O, Zipin-Roitman A, Mazor Y, Liefshitz B, Ben-Aroya S, Kupiec M. The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS One 2009; 4:e5497; http://dx.doi.org/10.1371/journal.pone.0005497; PMID: 19430531
- Banerjee S, Myung K. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot Cell 2004; 3:1557 - 66; http://dx.doi.org/10.1128/EC.3.6.1557-1566.2004; PMID: 15590829
- Smolikov S, Mazor Y, Krauskopf A. ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc Natl Acad Sci U S A 2004; 101:1656 - 61; http://dx.doi.org/10.1073/pnas.0307796100; PMID: 14745004
- Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci U S A 2003; 100:9906 - 11; http://dx.doi.org/10.1073/pnas.1633757100; PMID: 12909721
- Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol 2003; 13:1583 - 95; http://dx.doi.org/10.1016/S0960-9822(03)00578-5; PMID: 13678589
- Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J 2003; 22:4304 - 13; http://dx.doi.org/10.1093/emboj/cdg406; PMID: 12912927
- Huang M-E, Rio A-G, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci U S A 2003; 100:11529 - 34; http://dx.doi.org/10.1073/pnas.2035018100; PMID: 12972632
- Smith S, Hwang J-Y, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2004; 101:9039 - 44; http://dx.doi.org/10.1073/pnas.0403093101; PMID: 15184655
- Bell DW, Sikdar N, Lee KY, Price JC, Chatterjee R, Park H-D, et al, NISC Comparative Sequencing Program. Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet 2011; 7:e1002245; http://dx.doi.org/10.1371/journal.pgen.1002245; PMID: 21901109
- Sikdar N, Banerjee S, Lee KY, Wincovitch S, Pak E, Nakanishi K, et al. DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle 2009; 8:3199 - 207; http://dx.doi.org/10.4161/cc.8.19.9752; PMID: 19755857
- Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol Cell 2013; 50:273 - 80; http://dx.doi.org/10.1016/j.molcel.2013.02.012; PMID: 23499004
- Lee KY, Fu H, Aladjem MI, Myung K. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J Cell Biol 2013; 200:31 - 44; http://dx.doi.org/10.1083/jcb.201206084; PMID: 23277426
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007; 129:665 - 79; http://dx.doi.org/10.1016/j.cell.2007.05.003; PMID: 17512402
- Beattie TR, Bell SD. The role of the DNA sliding clamp in Okazaki fragment maturation in archaea and eukaryotes. Biochem Soc Trans 2011; 39:70 - 6; http://dx.doi.org/10.1042/BST0390070; PMID: 21265749
- Gomes XV, Burgers PM. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem 2001; 276:34768 - 75; http://dx.doi.org/10.1074/jbc.M011631200; PMID: 11432853
- Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 2004; 429:724 - 30; http://dx.doi.org/10.1038/nature02585; PMID: 15201901
- Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science 2011; 334:1675 - 80; http://dx.doi.org/10.1126/science.1211884; PMID: 22194570
- Hedglin M, Kumar R, Benkovic SJ. Replication clamps and clamp loaders. Cold Spring Harb Perspect Biol 2013; 5:a010165; http://dx.doi.org/10.1101/cshperspect.a010165; PMID: 23545418
- Yao NY, O’Donnell M. The RFC Clamp Loader: Structure and Function. Subcell Biochem 2012; 62:259 - 79; http://dx.doi.org/10.1007/978-94-007-4572-8_14; PMID: 22918590
- Kim J, MacNeill SA. Genome stability: a new member of the RFC family. Curr Biol 2003; 13:R873 - 5; http://dx.doi.org/10.1016/j.cub.2003.10.048; PMID: 14614842
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol 2000; 10:39 - 42; http://dx.doi.org/10.1016/S0960-9822(99)00263-8; PMID: 10660302
- Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci U S A 2001; 98:11236 - 41; http://dx.doi.org/10.1073/pnas.201373498; PMID: 11572977
- Majka J, Burgers PMJ. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci U S A 2003; 100:2249 - 54; http://dx.doi.org/10.1073/pnas.0437148100; PMID: 12604797
- Navadgi-Patil VM, Burgers PM. A tale of two tails: activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst) 2009; 8:996 - 1003; http://dx.doi.org/10.1016/j.dnarep.2009.03.011; PMID: 19464966
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 2001; 7:959 - 70; http://dx.doi.org/10.1016/S1097-2765(01)00254-4; PMID: 11389843
- Merkle CJ, Karnitz LM, Henry-Sánchez JT, Chen J. Cloning and characterization of hCTF18, hCTF8, and hDCC1. Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J Biol Chem 2003; 278:30051 - 6; http://dx.doi.org/10.1074/jbc.M211591200; PMID: 12766176
- Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 2001; 21:3144 - 58; http://dx.doi.org/10.1128/MCB.21.9.3144-3158.2001; PMID: 11287619
- Kubota T, Hiraga S, Yamada K, Lamond AI, Donaldson AD. Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol Cell Proteomics 2011; 10:M110:005561; http://dx.doi.org/10.1074/mcp.M110.005561; PMID: 21505101
- Hiraga S, Robertson ED, Donaldson AD. The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J 2006; 25:1505 - 14; http://dx.doi.org/10.1038/sj.emboj.7601038; PMID: 16525505
- Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol 2005; 25:5445 - 55; http://dx.doi.org/10.1128/MCB.25.13.5445-5455.2005; PMID: 15964801
- Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee C-G, Phillips B, et al. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc Natl Acad Sci U S A 1996; 93:12896 - 901; http://dx.doi.org/10.1073/pnas.93.23.12896; PMID: 8917516
- Yao N, Turner J, Kelman Z, Stukenberg PT, Dean F, Shechter D, et al. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells 1996; 1:101 - 13; http://dx.doi.org/10.1046/j.1365-2443.1996.07007.x; PMID: 9078370
- Yao NY, Johnson A, Bowman GD, Kuriyan J, O’Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J Biol Chem 2006; 281:17528 - 39; http://dx.doi.org/10.1074/jbc.M601273200; PMID: 16608854
- Shibahara K-i, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 1999; 96:575 - 85; http://dx.doi.org/10.1016/S0092-8674(00)80661-3; PMID: 10052459
- Parnas O, Zipin-Roitman A, Pfander B, Liefshitz B, Mazor Y, Ben-Aroya S, et al. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J 2010; 29:2611 - 22; http://dx.doi.org/10.1038/emboj.2010.128; PMID: 20571511
- Kubota T, Stead DA, Hiraga S-i, ten Have S, Donaldson AD. Quantitative proteomic analysis of yeast DNA replication proteins. Methods 2012; 57:196 - 202; http://dx.doi.org/10.1016/j.ymeth.2012.03.012; PMID: 22465796
- Parker JL, Bucceri A, Davies AA, Heidrich K, Windecker H, Ulrich HD. SUMO modification of PCNA is controlled by DNA. EMBO J 2008; 27:2422 - 31; http://dx.doi.org/10.1038/emboj.2008.162; PMID: 18701921
- Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J Cell Biol 2005; 171:947 - 54; http://dx.doi.org/10.1083/jcb.200508100; PMID: 16344309
- Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol 2006; 4:e366; http://dx.doi.org/10.1371/journal.pbio.0040366; PMID: 17105346
- Göhler T, Munoz IM, Rouse J, Blow JJ. PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair (Amst) 2008; 7:775 - 87; http://dx.doi.org/10.1016/j.dnarep.2008.02.001; PMID: 18353733
- Gali H, Juhasz S, Morocz M, Hajdu I, Fatyol K, Szukacsov V, et al. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res 2012; 40:6049 - 59; http://dx.doi.org/10.1093/nar/gks256; PMID: 22457066
- Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell 2012; 45:75 - 86; http://dx.doi.org/10.1016/j.molcel.2011.11.010; PMID: 22153967
- Lee KY, Yang K, Cohn MA, Sikdar N, D’Andrea AD, Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J Biol Chem 2010; 285:10362 - 9; http://dx.doi.org/10.1074/jbc.M109.092544; PMID: 20147293
- Yang K, Moldovan GL, Vinciguerra P, Murai J, Takeda S, D’Andrea AD. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev 2011; 25:1847 - 58; http://dx.doi.org/10.1101/gad.17020911; PMID: 21896657
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 1999; 9:27 - 43; PMID: 9927482
- Majka J, Chung BY, Burgers PMJ. Requirement for ATP by the DNA damage checkpoint clamp loader. J Biol Chem 2004; 279:20921 - 6; http://dx.doi.org/10.1074/jbc.M400898200; PMID: 15014082
- Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol 2000; 20:5888 - 96; http://dx.doi.org/10.1128/MCB.20.16.5888-5896.2000; PMID: 10913172
- Cai J, Yao N, Gibbs E, Finkelstein J, Phillips B, O’Donnell M, et al. ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits. Proc Natl Acad Sci U S A 1998; 95:11607 - 12; http://dx.doi.org/10.1073/pnas.95.20.11607; PMID: 9751713
- Podust VN, Tiwari N, Ott R, Fanning E. Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J Biol Chem 1998; 273:12935 - 42; http://dx.doi.org/10.1074/jbc.273.21.12935; PMID: 9582326
- Schmidt SLG, Gomes XV, Burgers PMJ. ATP utilization by yeast replication factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J Biol Chem 2001; 276:34784 - 91; http://dx.doi.org/10.1074/jbc.M011633200; PMID: 11432854
- Schmidt SLG, Pautz AL, Burgers PMJ. ATP utilization by yeast replication factor C. IV. RFC ATP-binding mutants show defects in DNA replication, DNA repair, and checkpoint regulation. J Biol Chem 2001; 276:34792 - 800; http://dx.doi.org/10.1074/jbc.M011633200; PMID: 11549622
- Davidson MB, Brown GW. The N- and C-termini of Elg1 contribute to the maintenance of genome stability. DNA Repair (Amst) 2008; 7:1221 - 32; http://dx.doi.org/10.1016/j.dnarep.2008.04.001; PMID: 18482875
- Symington LS. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res 1998; 26:5589 - 95; http://dx.doi.org/10.1093/nar/26.24.5589; PMID: 9837987
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 1999; 23:81 - 5; http://dx.doi.org/10.1038/12687; PMID: 10471504
- Saugar I, Parker JL, Zhao S, Ulrich HD. The genome maintenance factor Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res 2012; 40:245 - 57; http://dx.doi.org/10.1093/nar/gkr738; PMID: 21911365
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002; 419:135 - 41; http://dx.doi.org/10.1038/nature00991; PMID: 12226657
- Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci U S A 2005; 102:15954 - 9; http://dx.doi.org/10.1073/pnas.0504586102; PMID: 16247017
- Hishida T, Iwasaki H, Ohno T, Morishita T, Shinagawa H. A yeast gene, MGS1, encoding a DNA-dependent AAA(+) ATPase is required to maintain genome stability. Proc Natl Acad Sci U S A 2001; 98:8283 - 9; http://dx.doi.org/10.1073/pnas.121009098; PMID: 11459965
- Hishida T, Ohno T, Iwasaki H, Shinagawa H. Saccharomyces cerevisiae MGS1 is essential in strains deficient in the RAD6-dependent DNA damage tolerance pathway. EMBO J 2002; 21:2019 - 29; http://dx.doi.org/10.1093/emboj/21.8.2019; PMID: 11953321
- Branzei D, Seki M, Onoda F, Enomoto T. The product of Saccharomyces cerevisiae WHIP/MGS1, a gene related to replication factor C genes, interacts functionally with DNA polymerase Δ. Mol Genet Genomics 2002; 268:371 - 86; http://dx.doi.org/10.1007/s00438-002-0757-3; PMID: 12436259
- Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell 2004; 15:1736 - 45; http://dx.doi.org/10.1091/mbc.E03-08-0619; PMID: 14742714
- Warren CD, Eckley DM, Lee MS, Hanna JS, Hughes A, Peyser B, et al. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol Biol Cell 2004; 15:1724 - 35; http://dx.doi.org/10.1091/mbc.E03-09-0637; PMID: 14742710
- Rudra S, Skibbens RV. Sister chromatid cohesion establishment occurs in concert with lagging strand synthesis. Cell Cycle 2012; 11:2114 - 21; http://dx.doi.org/10.4161/cc.20547; PMID: 22592531
- Borges V, Smith DJ, Whitehouse I, Uhlmann F. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae. Chromosoma 2013; 122:121 - 34; http://dx.doi.org/10.1007/s00412-013-0396-y; PMID: 23334284
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 2009; 33:763 - 74; http://dx.doi.org/10.1016/j.molcel.2009.02.028; PMID: 19328069
- Zhang J, Shi X, Li Y, Kim B-J, Jia J, Huang Z, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 2008; 31:143 - 51; http://dx.doi.org/10.1016/j.molcel.2008.06.006; PMID: 18614053
- Ünal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science 2008; 321:566 - 9; http://dx.doi.org/10.1126/science.1157880; PMID: 18653894
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 2008; 321:563 - 6; http://dx.doi.org/10.1126/science.1157774; PMID: 18653893
- Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 2006; 23:723 - 32; http://dx.doi.org/10.1016/j.molcel.2006.07.007; PMID: 16934511
- Carson MJ, Hartwell L. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 1985; 42:249 - 57; http://dx.doi.org/10.1016/S0092-8674(85)80120-3; PMID: 3893744
- Adams AK, Holm C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol 1996; 16:4614 - 20; PMID: 8756617
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, et al. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet 2006; 2:e35; http://dx.doi.org/10.1371/journal.pgen.0020035; PMID: 16552446
- Parnas O, Amishay R, Liefshitz B, Zipin-Roitman A, Kupiec M. Elg1, the major subunit of an alternative RFC complex, interacts with SUMO-processing proteins. Cell Cycle 2011; 10:2894 - 903; http://dx.doi.org/10.4161/cc.10.17.16778; PMID: 21869594
- Hang LE, Liu X, Cheung I, Yang Y, Zhao X. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol 2011; 18:920 - 6; http://dx.doi.org/10.1038/nsmb.2100; PMID: 21743457
- Ferreira HC, Luke B, Schober H, Kalck V, Lingner J, Gasser SM. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol 2011; 13:867 - 74; http://dx.doi.org/10.1038/ncb2263; PMID: 21666682
- Almedawar S, Colomina N, Bermúdez-López M, Pociño-Merino I, Torres-Rosell J. A SUMO-dependent step during establishment of sister chromatid cohesion. Curr Biol 2012; 22:1576 - 81; http://dx.doi.org/10.1016/j.cub.2012.06.046; PMID: 22771040
- McAleenan A, Cordon-Preciado V, Clemente-Blanco A, Liu IC, Sen N, Leonard J, et al. SUMOylation of the α-kleisin subunit of cohesin is required for DNA damage-induced cohesion. Curr Biol 2012; 22:1564 - 75; http://dx.doi.org/10.1016/j.cub.2012.06.045; PMID: 22771042
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012; 151:807 - 20; http://dx.doi.org/10.1016/j.cell.2012.10.021; PMID: 23122649
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 2009; 16:314 - 20; http://dx.doi.org/10.1016/j.devcel.2009.01.001; PMID: 19217432