Abstract
The replication machinery, or the replisome, collides with a variety of obstacles during the normal process of DNA replication. In addition to damaged template DNA, numerous chromosome regions are considered to be difficult to replicate owing to the presence of DNA secondary structures and DNA-binding proteins. Under these conditions, the replication fork stalls, generating replication stress. Stalled forks are prone to collapse, posing serious threats to genomic integrity. It is generally thought that the replication checkpoint functions to stabilize the replisome and replication fork structure upon replication stress. This is important in order to allow DNA replication to resume once the problem is solved. However, our recent studies demonstrated that some replisome components undergo proteasome-dependent degradation during DNA replication in the fission yeast Schizosaccharomyces pombe. Our investigation has revealed the involvement of the SCFPof3 (Skp1-Cullin/Cdc53-F-box) ubiquitin ligase in replisome regulation. We also demonstrated that forced accumulation of the replisome components leads to abnormal DNA replication upon replication stress. Here we review these findings and present additional data indicating the importance of replisome degradation for DNA replication. Our studies suggest that cells activate an alternative pathway to degrade replisome components in order to preserve genomic integrity.
Introduction
The conditions for DNA replication are not ideal, owing to endogenous and exogenous replication stresses that lead to arrest of the replication fork. Arrested forks are among the most serious threats to genomic integrity, because they can break or rearrange, leading to genomic instability, a hallmark of cancer.Citation1-Citation4 To prevent such problems, cells possess a quality control system termed the DNA replication checkpoint. The loss of genomic integrity occurs at a high frequency in cells defective for the replication checkpoint pathway.Citation5-Citation9
Atop the replication checkpoint system stands phosphatidylinositol-3-kinase (PI3K)-related kinases such as human ATM (ataxia telangiectasia-mutated) and ATR (ATM and Rad3-related), fission yeast Rad3, and budding yeast Mec1 (). In the fission yeast Schizosaccharomyces pombe, in response to replication stress, Rad3 is recruited to ssDNA via interaction of its partner Rad26 (ATRIP in humans) with replication protein A (RPA). The Rad3-Rad26 complex works together with a trimeric checkpoint clamp 9-1-1 and 5-subunit clamp loader RFCRad17 (replication factor C-Rad17) to sense ssDNA accumulated at stalled replication forks and activate a signal transduction cascade. This signal is then mediated by the replication checkpoint mediator Mrc1, which channels the checkpoint signal to a downstream kinase Cds1, in order to arrest the cell cycle and facilitate DNA repair pathways ().Citation9-Citation12
Figure 1. Checkpoint activation and regulation of the replisome complex. (A) The replication checkpoint in fission yeast. Replication stress activates the Rad3-dependent checkpoint pathway, which is mediated by Mrc1 and the Swi1-Swi3 FPC. Rad3 sends a checkpoint signal to downstream checkpoint effector Cds1 to arrest the cell cycle and facilitate DNA repair pathways. Functional human homologs are indicated in parentheses. (B) SCFPof3-dependent replisome ubiquitination in response to replication stress. Natural impediments generate replication stress that activates SCFPof3 and ubiquitinates several replisome components for degradation. Replication stress also causes RPA accumulation, leading to checkpoint activation.
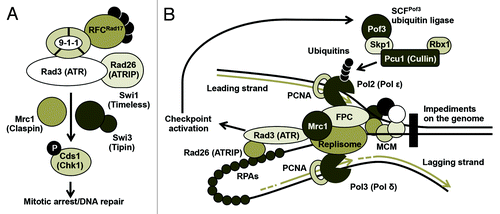
Another essential function of the replication checkpoint is to stabilize replication fork structure and the replisome complex in a replication-competent state.Citation13-Citation17 Studies found a group of proteins that are involved in fork stabilization. These proteins include the checkpoint mediator Mrc1 as well as Swi1 and Swi3 in S. pombe. Swi1 and Swi3 form the replication fork protection complex (FPC) and are required for efficient activation of Cds1 and for replication fork stability ().Citation18-Citation21 The functions of Mrc1 and the Swi1-Swi3 FPC are conserved among eukaryotes.Citation18,Citation21-Citation25 However, how these proteins protect replication forks and control replisome activities remains elusive.
It is generally thought that cells stabilize replisomes upon replication stress. However, it would also be advantageous to the cell to degrade the replisome and stop inappropriate replication when replisome progression is perturbed. It is also possible that some replisome components undergo degradation in order to complete DNA replication. Such mechanisms remain elusive. Interestingly, our recent investigation revealed that several replisome components are degraded in response to replication stress.Citation26 We have also showed evidence suggesting the role of proteasome-dependent replisome degradation in preventing genomic instability during DNA replication. Here we will discuss the mechanisms and physiological importance of replisome degradation in DNA replication.
Role of the FPC in Replisome Stabilization
Replication checkpoint studies often use chemical agents to stall replication forks. However, numerous chromosome regions present obstacles for DNA replication. These include replication fork blocking sites, DNA secondary structures caused by repeat sequences (such as telomeres), and DNA-binding proteins (such as the transcription machinery). These sites are difficult to replicate, causing replication fork arrest or breakage, resulting in replication stress during the normal course of DNA replication ().Citation3,Citation5,Citation21,Citation27-Citation29
Fission yeast Swi1 and its orthologs (Timeless in humans, Tof1 in budding yeast) are known to prevent DNA damage and hyperrecombination activity at natural obstacles scattered throughout the genome. These sites include rDNA pausing sites, repeat DNA sequences, highly transcribed loci, and telomeres, indicating that Swi1-related proteins are required for the smooth passage of replication forks and for the suppression of replication stress at difficult-to-replicate regions ().Citation21,Citation29-Citation38
To understand the role of the FPC in replisome stabilization, we recently examined the stability of major replisome components in the absence of Swi1, a subunit of the FPC.Citation26 Strikingly, major replicative DNA polymerases (Pol2 and Pol3) and MCM (minichromosome maintenance) helicase subunits (Mcm2/4/6) underwent degradation in swi1∆ cells, when cells were treated with cycloheximide, which inhibits new protein synthesis. In contrast, PCNA and Orc1, which are also essential for DNA replication, were stable.Citation26 These results suggest a role for Swi1 in preventing rapid degradation of replisome components. The degradation of DNA polymerases and helicases was inhibited when the proteasome was inactivated by the temperature-sensitive mts3–1 allele, which has a mutation in a proteasome subunit. Thus, the degradation of DNA polymerases and helicases is mediated by the ubiquitin-proteasome system. Indeed, Pol2 and Pol3 were ubiquitinated. Importantly, degradation of Pol2, Pol3, and Mcm4 was observed in chromatin-enriched fractions, suggesting that the chromatin fraction of these proteins undergoes degradation, in the absence of Swi1.Citation26 Furthermore, by using synchronized wild-type S. pombe cells, we reported that Pol2 degradation occurs during S phase. In contrast, there was no significant degradation of Pol3 and Mcm4 in wild-type cells, although they were unstable in swi1∆ cells. These results suggest that Pol2 is regulated by proteolysis during the normal course of the DNA replication process. Importantly, Pol2 degradation in S phase was further enhanced by swi1 deletion.Citation26 Considering that Swi1 is involved in suppression of DNA damage at difficult-to-replicate regions, which naturally exist in the genome,Citation21 our results may also suggest that cells promote a fast protein turnover of Pol2 when the fork encounters natural impediments.
Role of SCFPof3-dependent Pol2 Degradation in Faithful DNA Replication
Skp1-Cullin/Cdc53-F-box (SCF) ubiquitin ligases are often associated with protein degradation during S phase.Citation39 Consistently, we found that Pol2 degradation in wild-type cells was significantly inhibited in the absence of Skp1, a major component of SCF-type ubiquitin ligases.Citation40 The substrate specificity of SCF complexes is determined by the variable F-box subunit, a substrate-specific receptor.Citation41,Citation42 Among the many F-box proteins in S. pombe, we found that Pol2 degradation in both wild-type and swi1∆ cells was significantly inhibited in the absence of Pof3. Moreover, Pol2 was shown to co-purify with Pof3, suggesting the physiological importance of SCFPof3-dependent Pol2 degradation during DNA replication.Citation26
Interestingly, previous studies reported that Pof3 is involved in the preservation of genomic integrity.Citation43 SCFPof3 interacts with fission yeast Mcl1, a DNA polymerase α accessory factor related to budding yeast Ctf4,Citation43-Citation45 suggesting a role for Pof3 in DNA replication. In the budding yeast Saccharomyces cerevisiae, Dia2, a Pof3-related protein, also associates with the replisome and regulates replication forks.Citation46-Citation48 Genetic studies suggest that Dia2 is involved in regulating difficult-to-replicate regions of the genome.Citation49 Because there is a high chance of fork stalling and/or fork breakage at difficult-to-replicate regions, leading to replication stress and checkpoint activation,Citation21,Citation29 it is highly possible that Dia2 functions when the replisome encounters problems during replication. Indeed, Dia2 is stabilized when a checkpoint is activated, suggesting a role of Dia2/Pof3 in response to replication stress and during checkpoint activation ().Citation46,Citation49,Citation50 Furthermore, Dia2 is recruited to stalled forks and is involved in ubiquitination of Mrc1, a checkpoint mediator associated with the replisome.Citation46-Citation48 These findings suggest that Dia2/Pof3 regulates the replisome, in order to ensure proper DNA replication ().
pof3∆ cells showed an increased level of mitotic catastrophes in response to hydroxyurea (HU) and camptothecin (CPT). pof3∆ cells were sensitive to CPT and weakly sensitive to HU. pof3∆ cells also displayed a cell elongation phenotype, which is normally caused by accumulation of DNA damage or replication failure. Consistently, pof3∆ cells showed a delay in replication recovery after a low-dose CPT treatment, which did not affect replication recovery in wild-type cells.Citation26 Interestingly, Pol2 overexpression is deleterious to the cells (data not shown). Considering that pof3 deletion stabilizes Pol2 and other replisome components, our results suggest that SCFPof3-dependent replisome degradation plays an important role in preservation of genomic integrity during replication (). Interestingly, mitotic catastrophes and replication defects were further enhanced in swi1∆ pof3∆ double-deletion mutant cells. Considering that swi1 deletion causes replication stress at difficult-to-replicate regions, our results also suggest the importance of Pol2 degradation during replication of difficult-to-replicate regions.
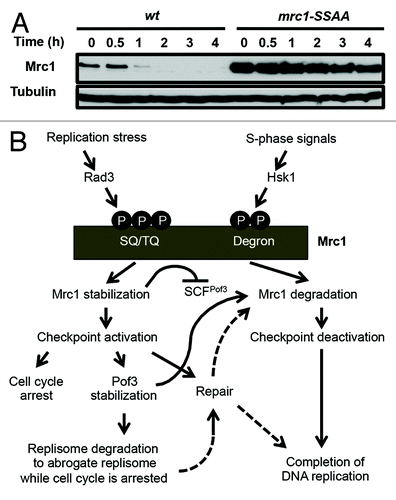
Figure 2. Regulation of Mrc1 cellular amounts during replication and checkpoint. (A) mrc1-S860A/S864A mutation stabilizes Mrc1. Exponentially growing cells of the indicated genotypes were treated with 0.1 mg/ml cycloheximide at 30 ˚C. Cellular amounts of Mrc1-FLAG and Mrc1-S860A/S864A-FLAG were examined at various time points between 0 and 4 h of cycloheximide treatment. The anti-FLAG (M2) antibody was used to detect Mrc1. Western blotting of tubulin was performed as a loading control. (B) Models of phosphorylation-dependent Mrc1 regulation during replication and checkpoint. Pof3 controls replisome quality via ubiquitination/degradation of Pol2 when the fork is adversely blocked and the checkpoint is activated, while Pof3 also regulates Mrc1 to deactivate checkpoint when the replisome is intact to resume and complete DNA replication.
Checkpoint programs are thought to stabilize replisomes upon replication stress.Citation1-Citation4 However, our results suggest that SCFPof3 promotes Pol2 degradation to abrogate replisome function in response to replication stress (). Such a mechanism may contribute to the prevention of inappropriate DNA replication that causes genomic instability.
Regulation of Mrc1 Degradation by SCFPof3 and Phosphorylation
In S. cerevisiae, Dia2 is involved in degradation of Mrc1, a replisome component required for the activation of the replication checkpoint kinase Rad53 (Cds1 ortholog).Citation25 Mrc1 also functions in coordination of polymerase and helicase functions at the fork.Citation51 As in the case of Dia2, we found that Pof3 is involved in Mrc1 degradation.Citation26 It is widely known that SCF-type ubiquitin ligases recognize phosphodegrons.Citation39 Interestingly, Mrc1 is hyperphosphorylated at the G1/S boundary and in response to replication-stressing agents.Citation52 This hyperphosphorylation is largely lost when the Hsk1 kinase is inactivated by a mutation in Hsk1, a Cdc7-related kinase essential for DNA replication,Citation52 suggesting the involvement of phosphorylation events in Mrc1 degradation. Indeed, fission yeast Mrc1 has a “phosphodegron” motif (DSGVGS859–864 aa), which can be recognized by an SCF ubiquitin ligase.Citation53 Strikingly, mutations in this motif (mrc1-S860A/S864A) significantly stabilized Mrc1 in fission yeast (). Furthermore, Mrc1 is also stabilized in hsk1–89 mutants.Citation53 Thus, Mrc1 is regulated by phosphorylation and degradation, in which Hsk1 and SCFPof3 have a key role.
Mrc1 hyperphosphorylation also depends on Rad3 and Tel1 kinases.Citation52,Citation54 Importantly, Rad3 and Tel1 phosphorylate SQ/TQ motifs within the SQ/TQ cluster of Mrc1 in response to replication stress,Citation54 while Hsk1-dependent phosphorylation occurs at other unknown sites.Citation52 Because Hsk1 has been shown to directly phosphorylate Mrc1,Citation52 it is possible that Hsk1 phosphorylates Mrc1 at its phosphodegron. Therefore, we suggest the involvement of a complex mechanism regulating Mrc1 phosphorylation and degradation. It is tempting to suggest that the balance between Rad3/Tel1-dependent and Hsk1-dependent phosphorylation events plays an important role in regulating replication and checkpoint processes (). It is straightforward to suggest that Mrc1 stabilization promotes Cds1-dependent checkpoint activation in response to replication stress. However, once the replication problem is solved, Mrc1 needs to be degraded in order to deactivate the checkpoint and complete DNA replication. We speculate that, to achieve this goal, Mrc1 undergoes dephosphorylation at Rad3/Tel1-dependent sites while preserving Hsk1-dependent phosphorylation, thus facilitating SCFPof3-dependent Mrc1 degradation and checkpoint deactivation ().
How SCFPof3 is Regulated During Replication and Checkpoint Processes
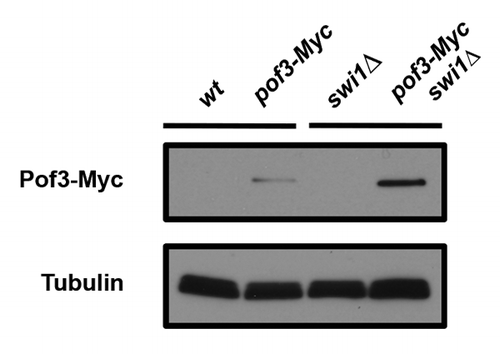
In S. cerevisiae, Dia2 levels are low in G1 and elevated during S phase. Dia2 itself is regulated by ubiquitination, and Dia2 degradation is inhibited in response to replication stress, such as HU treatment.Citation50 These results suggest that Dia2 activity is required in S phase and during checkpoint activation. Indeed, HU-dependent Dia2 stabilization is abrogated in checkpoint mutants, including chk1∆ and rad53-21 cells.Citation50 However, how Dia2 is regulated during checkpoint activation is largely unknown. Interestingly, we found in S. pombe that Pof3 levels are elevated in swi1∆ cells (). While Swi1 is involved in full activation of Cds1 (Rad53 ortholog), swi1∆ cells still possess a basal level of Cds1 activity.Citation55 In addition, swi1 deletion generates replication stress, which, in turn, activates the Chk1-dependent checkpoint pathway.Citation19,Citation31,Citation33,Citation35,Citation36,Citation55 Therefore, it is possible that SCFPof3 activity and/or levels are under the regulation of checkpoints ().
Figure 3. Pof3 is elevated in swi1∆ cells. Cells of the indicated genotypes were grown, and protein samples were prepared. Cellular amounts of Pof3-Myc were determined by using the anti-Myc (9E10) antibody. Tubulin levels were monitored as a loading control.
In budding yeast, in the absence of Dia2, an increased amount of Mrc1 leads to continuous activation of Rad53 after DNA damage, resulting in defects in checkpoint deactivation and in timely completion of DNA replication. These defects were partially suppressed by inactivation of Tof1 (Swi1 ortholog). This suppression is attributed to the fact that tof1∆ cells have lower Rad53 activity. Consistently, DNA damage sensitivity of dia2∆ cells was suppressed by tof1 deletion.Citation56 However, our data demonstrated in fission yeast that pof3∆ swi1∆ cells are more sensitive to HU and CPT than either single mutant,Citation26 indicating that further investigation of the role of Pof3-dependent Mrc1 degradation is needed.
Dia2 is stabilized when the checkpoint is activated, suggesting the role of Dia2/Pof3 in response to replication stress and during checkpoint activation.Citation46,Citation49,Citation50 In this model, Mrc1-dependent activation of the replication checkpoint kinase Rad53 (Cds1 ortholog) results in Dia2 stabilization.Citation57 However, studies also showed that Dia2 mediates ubiquitination/degradation of Mrc1, leading to deactivation of checkpoint, in order to resume and complete DNA replication.Citation47,Citation56 Therefore, Dia2/Pof3 appears to be involved in multiple steps during DNA replication and checkpoint processes. We suggest that Dia2/Pof3 controls replisome quality via ubiquitination/degradation of replisome components when the fork is adversely blocked and the checkpoint is activated, while Pof3 also regulates Mrc1 to deactivate checkpoint when the replisome is intact to resume and complete DNA replication ().
We proposed that SCFDia2/Pof3 is required to control DNA replication when the replication fork encounters natural barriers. Because chromosomes contain numerous barriers that are difficult to replicate, it will be important in the future to investigate whether Mrc1 degradation is precisely regulated at these genomic regions to control replication fork progression and activation of the replication checkpoint.
Concluding Remarks
We have made the interesting discovery that replisome components, including Pol2 and Mrc1, are highly unstable during replication.Citation26 Our data suggest the role of the SCFPof3 ubiquitin ligase in replisome regulation.Citation26 It is generally thought that checkpoint proteins stabilize replication forks and replisomes in response to replication stress.Citation1-Citation4 However, our findings suggest an alternative mechanism by which cells degrade the replisome when the fork is adversely blocked. This finding represents a paradigm shift in understanding the mechanisms of replisome maintenance. Therefore, our studies provide new mechanistic insights into replication stress response pathways. In addition, although a number of studies have focused on the processes of replication initiation and regulation of fork progression, how the replisome itself is regulated is largely unknown. Therefore, our findings will also fill the knowledge gap regarding the regulation of replisome components during the replication process.
| Abbreviations: | ||
| ATM | = | ataxia telangiectasia-mutated |
| ATR | = | ATM and Rad3-related |
| ATRIP | = | ATR interacting protein |
| CPT | = | camptothecin |
| FPC | = | fork protection complex |
| HU | = | hydroxyurea |
| MCM | = | minichromosome maintenance |
| Mrc1 | = | mediator of the replication checkpoint 1 |
| PCNA | = | proliferating cell nuclear antigen |
| PI3K | = | phosphatidylinositol-3-kinase |
| RFC | = | replication factor C |
| RPA | = | replication protein A |
| SCF | = | Skp1-Cullin/Cdc53-F-box |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs Katsunori Tanaka and Hisao Masai for S. pombe strains. This work was supported by the NIH (R01GM077604 to EN).
References
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 2010; 11:208 - 19; http://dx.doi.org/10.1038/nrm2852; PMID: 20177396
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 2006; 7:932 - 43; http://dx.doi.org/10.1038/nrm2058; PMID: 17139333
- Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J 2012; 443:13 - 26; http://dx.doi.org/10.1042/BJ20112100; PMID: 22417748
- Lambert S, Froget B, Carr AM. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst) 2007; 6:1042 - 61; http://dx.doi.org/10.1016/j.dnarep.2007.02.024; PMID: 17412649
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 2008; 9:204 - 17; http://dx.doi.org/10.1038/nrg2268; PMID: 18227811
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 2007; 19:238 - 45; http://dx.doi.org/10.1016/j.ceb.2007.02.009; PMID: 17303408
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 2008; 9:297 - 308; http://dx.doi.org/10.1038/nrm2351; PMID: 18285803
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 2008; 9:759 - 69; http://dx.doi.org/10.1038/nrm2514; PMID: 18813293
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007; 6:953 - 66; http://dx.doi.org/10.1016/j.dnarep.2007.02.015; PMID: 17531546
- Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci 2011; 36:133 - 40; http://dx.doi.org/10.1016/j.tibs.2010.09.005; PMID: 20947357
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet 2002; 36:617 - 56; http://dx.doi.org/10.1146/annurev.genet.36.060402.113540; PMID: 12429704
- Putnam CD, Jaehnig EJ, Kolodner RD. Perspectives on the DNA damage and replication checkpoint responses in Saccharomyces cerevisiae. DNA Repair (Amst) 2009; 8:974 - 82; http://dx.doi.org/10.1016/j.dnarep.2009.04.021; PMID: 19477695
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 2001; 412:557 - 61; http://dx.doi.org/10.1038/35087613; PMID: 11484058
- Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol 2001; 21:3913 - 25; http://dx.doi.org/10.1128/MCB.21.12.3913-3925.2001; PMID: 11359899
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002; 297:599 - 602; http://dx.doi.org/10.1126/science.1074023; PMID: 12142537
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 2001; 412:553 - 7; http://dx.doi.org/10.1038/35087607; PMID: 11484057
- Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell 2003; 11:1323 - 36; http://dx.doi.org/10.1016/S1097-2765(03)00169-2; PMID: 12769855
- McFarlane RJ, Mian S, Dalgaard JZ. The many facets of the Tim-Tipin protein families’ roles in chromosome biology. Cell Cycle 2010; 9:700 - 5; http://dx.doi.org/10.4161/cc.9.4.10676; PMID: 20139726
- Noguchi E, Noguchi C, McDonald WH, Yates JR 3rd, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 2004; 24:8342 - 55; http://dx.doi.org/10.1128/MCB.24.19.8342-8355.2004; PMID: 15367656
- Lee BS, Grewal SI, Klar AJ. Biochemical interactions between proteins and mat1 cis-acting sequences required for imprinting in fission yeast. Mol Cell Biol 2004; 24:9813 - 22; http://dx.doi.org/10.1128/MCB.24.22.9813-9822.2004; PMID: 15509785
- Leman AR, Noguchi E. Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle 2012; 11:3945 - 55; http://dx.doi.org/10.4161/cc.21989; PMID: 22987152
- Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol 2007; 366:36 - 52; http://dx.doi.org/10.1016/j.jmb.2006.10.097; PMID: 17141802
- Noguchi E. The DNA Replication Checkpoint and Preserving Genomic Integrity During DNA Synthesis. Nature Education 2010; 3:46
- Sabatinos SA. Replication Fork Stalling and the Fork Protection Complex. Nature Education 2010; 3:40
- Tanaka K. Multiple functions of the S-phase checkpoint mediator. Biosci Biotechnol Biochem 2010; 74:2367 - 73; http://dx.doi.org/10.1271/bbb.100583; PMID: 21150122
- Roseaulin LC, Noguchi C, Martinez E, Ziegler MA, Toda T, Noguchi E. Coordinated degradation of replisome components ensures genome stability upon replication stress in the absence of the replication fork protection complex. PLoS Genet 2013; 9:e1003213; http://dx.doi.org/10.1371/journal.pgen.1003213; PMID: 23349636
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 2007; 71:13 - 35; http://dx.doi.org/10.1128/MMBR.00030-06; PMID: 17347517
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 2005; 6:729 - 42; http://dx.doi.org/10.1038/nrg1689; PMID: 16205713
- Leman AR, Noguchi E. The replication fork: Understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes (Basel) 2013; 4:1 - 32; http://dx.doi.org/10.3390/genes4010001; PMID: 23599899
- Cherng N, Shishkin AA, Schlager LI, Tuck RH, Sloan L, Matera R, et al. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc Natl Acad Sci U S A 2011; 108:2843 - 8; http://dx.doi.org/10.1073/pnas.1009409108; PMID: 21282659
- Leman AR, Dheekollu J, Deng Z, Lee SW, Das MM, Lieberman PM, et al. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle 2012; 11:2337 - 47; http://dx.doi.org/10.4161/cc.20810; PMID: 22672906
- Liu G, Chen X, Gao Y, Lewis T, Barthelemy J, Leffak M. Altered replication in human cells promotes DMPK (CTG)(n) · (CAG)(n) repeat instability. Mol Cell Biol 2012; 32:1618 - 32; http://dx.doi.org/10.1128/MCB.06727-11; PMID: 22354993
- Pryce DW, Ramayah S, Jaendling A, McFarlane RJ. Recombination at DNA replication fork barriers is not universal and is differentially regulated by Swi1. Proc Natl Acad Sci U S A 2009; 106:4770 - 5; http://dx.doi.org/10.1073/pnas.0807739106; PMID: 19273851
- Razidlo DF, Lahue RS. Mrc1, Tof1 and Csm3 inhibit CAG.CTG repeat instability by at least two mechanisms. DNA Repair (Amst) 2008; 7:633 - 40; http://dx.doi.org/10.1016/j.dnarep.2008.01.009; PMID: 18321795
- Rozenzhak S, Mejía-Ramírez E, Williams JS, Schaffer L, Hammond JA, Head SR, et al. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet 2010; 6:e1001032; http://dx.doi.org/10.1371/journal.pgen.1001032; PMID: 20661445
- Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev 2012; 26:581 - 93; http://dx.doi.org/10.1101/gad.184697.111; PMID: 22426534
- Shishkin AA, Voineagu I, Matera R, Cherng N, Chernet BT, Krasilnikova MM, et al. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol Cell 2009; 35:82 - 92; http://dx.doi.org/10.1016/j.molcel.2009.06.017; PMID: 19595718
- Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci U S A 2008; 105:9936 - 41; http://dx.doi.org/10.1073/pnas.0804510105; PMID: 18632578
- Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 2013; 14:369 - 81; http://dx.doi.org/10.1038/nrm3582; PMID: 23657496
- Lehmann A, Katayama S, Harrison C, Dhut S, Kitamura K, McDonald N, et al. Molecular interactions of fission yeast Skp1 and its role in the DNA damage checkpoint. Genes Cells 2004; 9:367 - 82; http://dx.doi.org/10.1111/j.1356-9597.2004.00730.x; PMID: 15147268
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol 2004; 5:739 - 51; http://dx.doi.org/10.1038/nrm1471; PMID: 15340381
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 2006; 6:369 - 81; http://dx.doi.org/10.1038/nrc1881; PMID: 16633365
- Katayama S, Kitamura K, Lehmann A, Nikaido O, Toda T. Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol Biol Cell 2002; 13:211 - 24; http://dx.doi.org/10.1091/mbc.01-07-0333; PMID: 11809834
- Mamnun YM, Katayama S, Toda T. Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochem Biophys Res Commun 2006; 350:125 - 30; http://dx.doi.org/10.1016/j.bbrc.2006.09.024; PMID: 16997270
- Williams DR, McIntosh JR. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell 2002; 1:758 - 73; http://dx.doi.org/10.1128/EC.1.5.758-773.2002; PMID: 12455694
- Koepp DM, Kile AC, Swaminathan S, Rodriguez-Rivera V. The F-box protein Dia2 regulates DNA replication. Mol Biol Cell 2006; 17:1540 - 8; http://dx.doi.org/10.1091/mbc.E05-09-0884; PMID: 16421250
- Mimura S, Komata M, Kishi T, Shirahige K, Kamura T. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J 2009; 28:3693 - 705; http://dx.doi.org/10.1038/emboj.2009.320; PMID: 19910927
- Morohashi H, Maculins T, Labib K. The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr Biol 2009; 19:1943 - 9; http://dx.doi.org/10.1016/j.cub.2009.09.062; PMID: 19913425
- Blake D, Luke B, Kanellis P, Jorgensen P, Goh T, Penfold S, et al. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae.. Genetics 2006; 174:1709 - 27; http://dx.doi.org/10.1534/genetics.106.057836; PMID: 16751663
- Kile AC, Koepp DM. Activation of the S-phase checkpoint inhibits degradation of the F-box protein Dia2. Mol Cell Biol 2010; 30:160 - 71; http://dx.doi.org/10.1128/MCB.00612-09; PMID: 19858292
- Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell 2008; 32:106 - 17; http://dx.doi.org/10.1016/j.molcel.2008.08.020; PMID: 18851837
- Matsumoto S, Shimmoto M, Kakusho N, Yokoyama M, Kanoh Y, Hayano M, et al. Hsk1 kinase and Cdc45 regulate replication stress-induced checkpoint responses in fission yeast. Cell Cycle 2010; 9:4627 - 37; http://dx.doi.org/10.4161/cc.9.23.13937; PMID: 21099360
- Shimmoto M, Matsumoto S, Odagiri Y, Noguchi E, Russell P, Masai H. Interactions between Swi1-Swi3, Mrc1 and S phase kinase, Hsk1 may regulate cellular responses to stalled replication forks in fission yeast. Genes Cells 2009; 14:669 - 82; http://dx.doi.org/10.1111/j.1365-2443.2009.01300.x; PMID: 19422421
- Zhao H, Tanaka K, Nogochi E, Nogochi C, Russell P. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol Cell Biol 2003; 23:8395 - 403; http://dx.doi.org/10.1128/MCB.23.22.8395-8403.2003; PMID: 14585996
- Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol 2003; 23:7861 - 74; http://dx.doi.org/10.1128/MCB.23.21.7861-7874.2003; PMID: 14560029
- Fong CM, Arumugam A, Koepp DM. The Saccharomyces cerevisiae F-box protein Dia2 is a Mediator ofS-phase Checkpoint Recovery from DNA Damage. Genetics 2013; 193:483 - 99; http://dx.doi.org/10.1534/genetics.112.146373; PMID: 23172854
- Koepp DM. The replication stress response and the ubiquitin system: a new link in maintaining genomic integrity. Cell Div 2010; 5:8; http://dx.doi.org/10.1186/1747-1028-5-8; PMID: 20219119