The functional diversity of the proteome is enhanced by dynamic post-translational modifications (PTMs), and it is now clear that the cell exploits a wide range of these to enable the coordination of cellular responses. One such modification is methylation of arginine residues catalyzed by protein arginine methyltransferases (PRMTs). Arginine methylation modifies many proteins but has been best characterized in the context of gene expression. In a manner analogous to lysine methylation, arginine methylation of histone tails either positively or negatively regulates gene expression through the modulation of chromatin-bound complexes. This mechanism of transcriptional regulation may be relevant in cancer, as many transcription factors and epigenetic regulators are overexpressed or mutated in human tumors.
A classic example of oncogenic transcriptional regulation is the AP-1 family member c-Jun, which drives abnormal cell proliferation and transformation by cooperating with various oncogenes and carcinogens.Citation1 c-Jun-mediated transactivation of a specific set of genes requires the regulated recruitment of coactivators or repressors. In line with this, we recently identified RACO-1 as a c-Jun coactivator that links oncogenic growth factor signaling to AP-1 gene transactivationCitation2. In this new study, we further characterize the molecular regulation of RACO-1 and identify a role for arginine methylation in the AP-1 response.Citation3 We were able to show that PRMT1 specifically methylates RACO-1 on two arginine residues (R98 and R109), and that this methylation event enables RACO-1 to adopt a conformation permissive for c-Jun binding (). Consequently, depletion of PRMT1 by shRNA reduces RACO-1 methylation and c-Jun binding, leading to changes in gene expression similar to those caused by c-Jun depletion. These findings strongly imply that PRMT1, RACO-1, and c-Jun cooperate in controlling transcription of the same genes, and that methylation brings about a series of events that modulates AP-1 transactivation.
Figure 1. Proposed model for the effects of PRMT1-mediated arginine methylation in the regulation of RACO-1 stability and c-Jun binding. Demethylated RACO-1 (top left) is unstable due to autoubiquitylation targeting lysine residues within the C-terminal domain of the protein. Methylation of arginine 98 and arginine 109 (shown as the gray box) induces a conformational change enabling an intramolecular interaction between the N-terminal and C-terminal portions of the protein. Autoubiquitylation is subsequently inhibited and K63-linked ubiquitylation induced, leading to protein stabilization. Once stabilized by methylation, RACO-1 forms a dimeric complex via the C-terminal dimerization domain of each monomer (blue oval shape). Dimeric RACO-1 can subsequently interact with c-Jun and drive AP-1 gene activation. K, lysine residues; R, arginine methylation (Me) sites (98 and 109); U, ubiquitin.
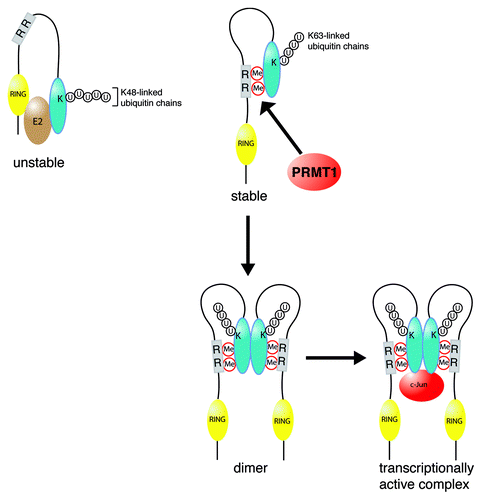
Arginine methylation, like other PTMs, modulates cellular behavior through promoting or preventing protein-protein interactions. The fact that nuclear but not cytoplasmic RACO-1 interacts with c-Jun implied that nuclear pools of RACO-1 adopted a specific conformation enabling c-Jun binding. We discovered that the regulation of RACO-1 appears to involve both intra- and intermolecular interactions. First, RACO-1 can dimerize, and dimeric rather than monomeric RACO-1 preferentially binds to c-Jun. We mapped the dimerization domain to the C-terminal tail (, domain shown in blue), but found that a second N-terminal fragment containing the two methylarginine residues also bound to the dimerization domain (, domain shown in gray). PRMT1-mediated methylation of R98 and R109 was pivotal to this N-to-C-terminal cis-interaction and subsequently required for dimerization. Hence we have a scenario where arginine methylation appears to be the critical catalyst that facilitates RACO-1 conformational changes, including a N-to-C-terminal cis- and a C-terminal tail-to-tail trans-interaction. Together, this ultimately enables RACO-1 to function as a c-Jun coactivator.
These findings raise a number of interesting points. First, methylation of RACO-1 somehow promotes a conformational change that enables RACO-1 to adopt a dimeric conformation that enables c-Jun binding. Mechanistically how this occurs is unknown; however, methylation does change the overall shape of the arginine guanidino group and reduces the number of potential hydrogen bonds that can be formed. Hence, these biochemical alterations have the potential to modulate binding interactions that occur between protein partners.
A second intriguing observation we made was that expression of PRMT1 stabilizes RACO-1. Previously we had noticed that activation of MAP kinase pathways leads to a switch from K48-linked to K63-linked ubiquitin chain formation, which prevents proteasomal degradation of RACO-1Citation2. In this latest study, we showed that arginine methylation is a prerequisite for this event, because mutation of the methyl-acceptor site or depletion of PRMT1 substantially suppressed K63-linked ubiquitin chain formation and thus RACO-1 stability. These findings add to the growing number of proteins involved in gene regulation that are subject to multiple forms of PTM in a co-operative or mutually exclusive manner. For example, histone H3R4 methylation by PRMT1 is required for H3/H4 acetylation,Citation4 while methylation of FOXO1 blocks Akt-mediated phosphorylation, preventing stress-induced apoptosis.Citation5
Arginine methylation is highly correlated with cancer development: dimethylation of histone H4R3 positively correlates with increased tumor grade in prostate cancer,Citation6 ERα is methylated in breast biopsies, and the interaction of PRMT1 with the AML1–ETO fusion protein promotes leukemic cell proliferation.Citation7 We know that RACO-1 genetically cooperates with APCmin mutation and oncogenic Ras in intestinal cancer development,Citation2 and that PRMT1 expression levels tend to be elevated in colorectal cancers with a poor prognosis.Citation8 Hence methylation of RACO-1 could be a potential driver of intestinal cancer.
Other chromatin bound non-histone proteins are also regulated by PRMT1, including the nuclear receptor cofactors RIP140 and PGC-1α8, both of which alter transcription and have been implicated in cancer. Together with these studies, our findings highlight a growing trend demonstrating the importance of PRMT1-mediated methylation in gene expression and cancer. Inhibiting PRMT1 activity or modulating the changes in protein complex formation brought about by methylarginine could therefore offer an attractive alternative strategy for rational drug design in cancer therapy.
References
- Shaulian E. AP-1--The Jun proteins: Oncogenes or tumor suppressors in disguise?. Cell Signal 2010; 22:894 - 9; http://dx.doi.org/10.1016/j.cellsig.2009.12.008; PMID: 20060892
- Davies CC, Chakraborty A, Cipriani F, Haigh K, Haigh JJ, Behrens A. Identification of a co-activator that links growth factor signalling to c-Jun/AP-1 activation. Nat Cell Biol 2010; 12:963 - 72; http://dx.doi.org/10.1038/ncb2098; PMID: 20852630
- Davies CC, Chakraborty A, Diefenbacher ME, Skehel M, Behrens A. Arginine methylation of the c-Jun coactivator RACO-1 is required for c-Jun/AP-1 activation. EMBO J 2013; 32:1556 - 67; http://dx.doi.org/10.1038/emboj.2013.98; PMID: 23624934
- Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev 2005; 19:1885 - 93; http://dx.doi.org/10.1101/gad.1333905; PMID: 16103216
- Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell 2008; 32:221 - 31; http://dx.doi.org/10.1016/j.molcel.2008.09.013; PMID: 18951090
- Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, et al. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell 2008; 31:212 - 21; http://dx.doi.org/10.1016/j.molcel.2008.05.025; PMID: 18657504
- Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer 2013; 13:37 - 50; http://dx.doi.org/10.1038/nrc3409; PMID: 23235912
- Mathioudaki K, Papadokostopoulou A, Scorilas A, Xynopoulos D, Agnanti N, Talieri M. The PRMT1 gene expression pattern in colon cancer. Br J Cancer 2008; 99:2094 - 9; http://dx.doi.org/10.1038/sj.bjc.6604807; PMID: 19078953