Organization of eukaryotic genomes into compact, nucleoprotein fibers is key for stuffing a lot of DNA into a tiny nucleus, but these structures also create a requirement for machineries that promote DNA accessibility for nuclear processes such as gene transcription, DNA repair, recombination, and replication. Arguably the most notable of such machineries is a large family of ATP-dependent chromatin-remodeling enzymes.Citation1 These are typically multi-subunit enzymes that perform the “heavy lifting” of chromosome dynamics, as they can use the energy from ATP hydrolysis to unfold chromatin fibers, mobilize nucleosomes within the fiber, evict histone components of the nucleosome, or catalyze incorporation or removal of variant histones that have distinct biochemical properties. These enzymes harbor an ATPase subunit that is related to the SF2 superfamily of DNA helicases, and although these enzymes lack helicase activity, several enzymes have ATP-dependent DNA translocase activity that is likely central for their remodeling activities.
As one might expect, these potent chromatin-remodeling enzymes are tightly regulated within cells, and four basic strategies have emergedCitation2,Citation3: (1) direct interaction with sequence-specific DNA binding proteins can target their activity to specific loci; (2) posttranslational modification of remodeling enzymes can modulate enzymatic activity; (3) binding of non-histone proteins to nucleosomal arrays can block remodeling activity and function as molecular “shields”; and (4) histone modifications can regulate remodeling. In most cases, these mechanisms serve to restrain remodeling activity, but our recent work uncovered a novel role for histone acetylation in altering the substrate specificity and overall activity of the SWR-C and INO80 remodeling enzymes.Citation4
Yeast SWR-C and INO80 are members of a subfamily of chromatin-remodeling enzymes conserved in all eukaryotes.Citation5 A crucial role for these multi-subunit enzymes is to control chromosomal distribution of the H2A.Z histone variant that marks nucleosomes at promoters of protein-coding genes, replication origins, centromeres, and chromatin boundary elements. SWR-C is essential for H2A.Z deposition. It evicts H2A/H2B dimers from a nucleosome and replaces them with H2A.Z/H2B dimers in an ATP-dependent reaction. In contrast, INO80 catalyzes the reverse reaction, removing H2A.Z and replacing it with H2A.Citation6 Recently, we discovered that acetylation of histone H3-K56 disrupts the substrate specificity of SWR-C, allowing it to perform like a combination of SWR-C and INO80, whereby it can catalyze deposition of either H2A.Z or H2A into nucleosomal substrates.Citation4 These studies suggested a model where H3-K56Ac promotes cycles of continuous dimer exchange that lowers H2A.Z levels at promoters and stimulates nucleosome turnover ().
Figure 1. Regulatory interaction between SWR-C and H3K56Ac mediates multiple cycles of histone-exchange. While SWR-C only catalyzes H2A.Z deposition on unacetylated nucleosomes, SWR-C can catalyze both deposition and eviction of H2A.Z on H3-K56Ac nucleosomes, leading to high turnover of H2A.Z.
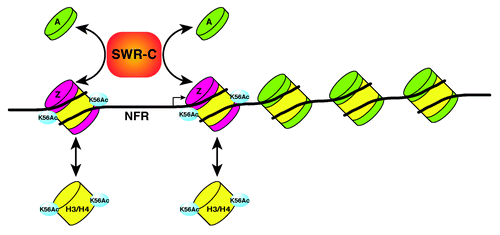
How does acetylation of a single histone lysine regulate SWR-C substrate selectivity? When SWR-C catalyzes replacement of H2A/H2B with H2A.Z/H2B dimers, the Swc2 subunit of SWR-C may bind the H2A.Z nucleosome and prevent activation of the Swr1 ATPase. In the absence of Swc2, the inhibition of Swr1 is relieved, the H2A.Z nucleosome is aberrantly recognized as a substrate, and a second round of dimer exchange ensues.Citation4 Thus, SWR-C functions much like a machine that can recognize reaction intermediates and steer them to the final product. And in this context, H3-K56Ac functions as a regulatory switch that alters the reaction cycle by directly, or indirectly, impacting Swc2 function.
Swc2 is the primary H2A.Z binding subunit within SWR-C, and it shares homology with mammalian YL-1, a subunit of related Srcap and Tip60 complexes. Interestingly, yeast INO80 also contains a subunit with homology to YL-1, called Ies6, which is essential for INO80 function.Citation7 It seems likely that Ies6 might also control dimer exchange activities of INO80, much like the role of Swc2 within SWR-C. Since H3-K56Ac also stimulates the dimer exchange activity of INO80, perhaps this histone mark also impinges on Ies6 functions. Whereas Swc2 and YL-1 share an abundance of both basic and acidic residues within their N-terminal regions, Ies6 lacks the acidic region. This suggests a simple model where Swc2 interacts with both DNA and histones, and Ies6 may play a more general role in coupling INO80 activity to nucleosomal DNA.
The acetylation of H3-K56 occurs on free, newly synthesized histones, and thus this mark is abundant within newly replicated chromatin.Citation8 H3-K56Ac stabilizes stalled replication forks by an unknown mechanism, but persistent H3-K56Ac is also deleterious for genome stability. Consequently, this mark is removed from bulk chromatin by the Hst3 and Hst4 deacetylases at the end of S phase. Whether these enzymes also regulate levels of H3-K56Ac in interphase is not yet known. Interestingly, SWR-C is also required for growth in conditions of replication stress, suggesting that H2A.Z might also play a role during S phase. Furthermore, the combination of H2A.Z deposition behind the fork and high levels of H3-K56ac on the new, daughter nucleosomes might generate a cell cycle window with highly dynamic nucleosomes. This dynamic state might be key for the stability of stalled forks, or it might facilitate other fork-associated events, such as DNA mismatch repair. Given the key role of H2A.Z in nearly every nuclear process, and the abundance of H3-K56Ac, it seems likely that there will be many examples where this regulatory interaction is exploited.
References
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009; 78:273 - 304; http://dx.doi.org/10.1146/annurev.biochem.77.062706.153223; PMID: 19355820
- Peterson CL. Chromatin remodeling enzymes: taming the machines. Third in review series on chromatin dynamics. EMBO Rep 2002; 3:319 - 22; http://dx.doi.org/10.1093/embo-reports/kvf075; PMID: 11943761
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311:844 - 7; http://dx.doi.org/10.1126/science.1124000; PMID: 16469925
- Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 2013; 340:195 - 9; http://dx.doi.org/10.1126/science.1229758; PMID: 23580526
- Watanabe S, Peterson CL. The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harb Symp Quant Biol 2010; 75:35 - 42; http://dx.doi.org/10.1101/sqb.2010.75.063; PMID: 21502417
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011; 144:200 - 13; http://dx.doi.org/10.1016/j.cell.2010.12.021; PMID: 21241891
- Chambers AL, Ormerod G, Durley SC, Sing TL, Brown GW, Kent NA, et al. The INO80 chromatin remodeling complex prevents polyploidy and maintains normal chromatin structure at centromeres. Genes Dev 2012; 26:2590 - 603; http://dx.doi.org/10.1101/gad.199976.112; PMID: 23207916
- Peterson CL. Genome integrity: a HAT needs a chaperone. Curr Biol 2007; 17:R324 - 6; http://dx.doi.org/10.1016/j.cub.2007.03.011; PMID: 17470345