The adenovirus E4 open reading frame 4 (E4orf4) protein is a multifunctional viral regulator that contributes to temporal regulation of the progression of viral infection. When expressed outside the context of the virus, E4orf4 induces p53-independent cell death in transformed cells. Oncogenic transformation of primary cells in tissue culture sensitizes them to cell killing by E4orf4,Citation1 indicating that E4orf4 research may have implications for cancer therapy. It has been further reported that E4orf4 induces a caspase-independent, non-classical apoptotic pathway that maintains crosstalk with classical caspase-dependent pathways.Citation2,Citation3 An investigation into the mechanisms involved in E4orf4-induced cell death revealed that E4orf4 interacts with the heterotrimeric protein phosphatase 2A (PP2A) through direct association with its regulatory B subunits, and the interaction mediated by the PP2A Bα/B55 subunit is required for inducing cell death.Citation1 Furthermore, E4orf4 recruits PP2A to a new substrate, the ACF chromatin remodeling factor, which contributes to E4orf4 functions.Citation4 E4orf4 has also been reported to associate with members of the Src kinase family, leading to its Tyr phosphorylation and to deregulation of Src signaling, resulting in enhanced cell death.Citation2 We showed previously that the interaction between E4orf4 and PP2A and its toxic consequences were conserved from yeast to mammals,Citation5 indicating a high degree of evolutionary conservation of the underlying mechanisms. This finding suggested the feasibility of using various model organisms for studying E4orf4-induced cell death. Indeed, our work in yeast revealed a novel E4orf4 partner, Ynd1, a Golgi UDPase that contributes to E4orf4 toxicity.Citation6
Recently we utilized Drosophila melanogaster as a model system to obtain new insights into the mechanisms underlying E4orf4-induced cell death in a whole organism.Citation7 This system was chosen for its excellent genetic tools and because of the availability of a wealth of public stock centers and databases, potentially facilitating efficient research of the E4orf4 network.
The Drosophila system was determined to be suitable for E4orf4 research by examining the similarity between E4orf4-induced cell death in mammalian cells and in the fly. We found that expression of E4orf4 in fly eyes or wings led to minor tissue damage including a small rough eye and disappearance of some wing substructures. Generating the E4orf4-induced phenotypes required both PP2A-B55 and Src, demonstrating that the functional interaction between E4orf4 and these partners was conserved. Furthermore, expression of the caspase inhibitors dIAP1 and p35 only partially reduced the E4orf4 effects, indicating that E4orf4-induced cell death in the fly relied on both caspase-dependent and -independent mechanisms, as was shown in mammalian cells in tissue culture. However, caspase dependence appeared to be more prominent in flies.Citation3,Citation7 We concluded that the interaction between E4orf4 and its partners, PP2A and Src, which initiates the death process, is similar in mammalian cells and in flies, but the crosstalk between the upstream caspase-independent part of the pathway and downstream classical apoptotic pathways may be more pronounced in the fly.
Once we established that E4orf4-induced cell death was very well conserved from flies to mammals, we continued using Drosophila to investigate the underlying mechanisms. We examined E4orf4-expressing cells in wing and eye imaginal discs and observed that only a fraction of these cells contained the active effector caspase, caspase-3. Furthermore, E4orf4-expressing cells containing active caspase-3 in the wing disc were extruded from the living tissue in a manner similar to previously described “undead” cells obtained by inducing apoptosis while simultaneously inhibiting caspase activation. This observation guided us toward testing whether E4orf4 could both activate and inhibit cell death. Indeed, we found that E4orf4 expression resulted in reduced severity of phenotypes induced by the pro-apoptotic genes reaper, hid, or grim (RHG). Furthermore, E4orf4 inhibited cell death induced by JNK activation in the fly, and JNK inhibition did not reduce E4orf4-induced cell death, and even enhanced it.Citation7 Thus our results indicated that E4orf4 activated a unique mode of programmed cell death, which differs from classical apoptotic pathways induced by RHG genes or JNK signaling. Moreover, the results provided new insight into E4orf4 activity, revealing that it can both activate and inhibit cell death in normal Drosophila tissues. This finding evokes a possible explanation for our previous observation showing that E4orf4 was more toxic in cancer cells than in normal cells in tissue culture.Citation1 We hypothesize that while cell death is inhibited by E4orf4 in normal cells, this may not occur in cancer cells, allowing E4orf4 to kill cancer cells more efficiently.
Inhibition of cell death by E4orf4 is also consistent with a protective role for this viral protein during adenovirus infection, which was suggested by previous experiments showing that a mutant adenovirus lacking E4orf4 was more cytotoxic in untransformed rodent cells. Preventing premature cell death is beneficial to the virus, as it allows the infection to progress and culminate successfully.
In summary, our studies of E4orf4 in Drosophila highlight the great degree of conservation of the mechanisms underlying E4orf4 functions and provide new insight explaining why E4orf4 kills normal cells inefficiently. It remains to be investigated whether the more effective cell killing induced by E4orf4 in cancer cells stems from loss of the E4orf4 ability to inhibit apoptosis in these cells. ()
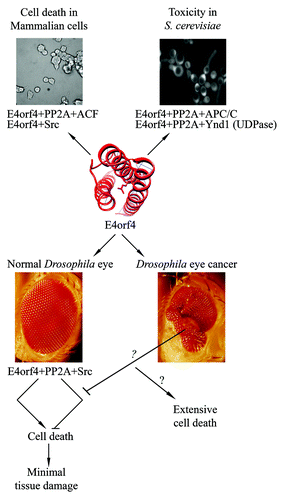
Figure 1. Cell killing by E4orf4 and its partners is conserved in evolution and is accompanied by inhibition of classical apoptosis in normal Drosophila tissues. Mechanisms underlying E4orf4-induced cell death were investigated in various organisms. Work in mammalian cells in tissue culture revealed a role for PP2A, Src kinases, and the ACF chromatin remodeling factor in E4orf4-induced cell death.Citation1,Citation2,Citation4 Work in yeast revealed the roles of PP2A, Golgi UDPase (Ynd1), and the anaphase-promoting-complex/cyclosome (APC/C) in E4orf4 toxicity.Citation5,Citation6 Work in Drosophila revealed that E4orf4 induced PP2A- and Src-dependent cell death in normal tissues while inhibiting classical apoptosis.Citation7 The concomitant induction and inhibition of cell death resulted in minor damage to normal tissues. We hypothesize that the more effective cell killing induced by E4orf4 in cancer cells may stem from reduced inhibition of classical apoptosis in these cells. This as-yet-untested hypothesis is represented by question marks. E4orf4 is represented by its structural model.Citation8
References
- Shtrichman R, Sharf R, Barr H, Dobner T, Kleinberger T. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc Natl Acad Sci U S A 1999; 96:10080 - 5; http://dx.doi.org/10.1073/pnas.96.18.10080; PMID: 10468565
- Lavoie JN, Champagne C, Gingras M-C, Robert A. Adenovirus E4 open reading frame 4-induced apoptosis involves dysregulation of Src family kinases. J Cell Biol 2000; 150:1037 - 56; http://dx.doi.org/10.1083/jcb.150.5.1037; PMID: 10973994
- Livne A, Shtrichman R, Kleinberger T. Caspase activation by adenovirus E4orf4 protein is cell line specific and is mediated by the death receptor pathway. J Virol 2001; 75:789 - 98; http://dx.doi.org/10.1128/JVI.75.2.789-798.2001; PMID: 11134292
- Brestovitsky A, Sharf R, Mittelman K, Kleinberger T. The adenovirus E4orf4 protein targets PP2A to the ACF chromatin-remodeling factor and induces cell death through regulation of SNF2h-containing complexes. Nucleic Acids Res 2011; 39:6414 - 27; http://dx.doi.org/10.1093/nar/gkr231; PMID: 21546548
- Kornitzer D, Sharf R, Kleinberger T. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J Cell Biol 2001; 154:331 - 44; http://dx.doi.org/10.1083/jcb.200104104; PMID: 11470822
- Maoz T, Koren R, Ben-Ari I, Kleinberger T. YND1 interacts with CDC55 and is a novel mediator of E4orf4-induced toxicity. J Biol Chem 2005; 280:41270 - 7; http://dx.doi.org/10.1074/jbc.M507281200; PMID: 16227198
- Pechkovsky A, Lahav M, Bitman E, Salzberg A, Kleinberger T. E4orf4 induces PP2A- and Src-dependent cell death in Drosophila melanogaster and at the same time inhibits classic apoptosis pathways. Proc Natl Acad Sci U S A 2013; 110:E1724 - 33; http://dx.doi.org/10.1073/pnas.1220282110; PMID: 23613593
- Horowitz B, Sharf R, Avital-Shacham M, Pechkovsky A, Kleinberger T. Structure- and modeling-based identification of the adenovirus E4orf4 binding site in the protein phosphatase 2A B55α subunit. J Biol Chem 2013; 288:13718 - 27; http://dx.doi.org/10.1074/jbc.M112.343756; PMID: 23530045