Helicases are widespread enzymes that share a core RecA fold and characteristic amino acid motifs which together form an ATP binding/hydrolysis site.Citation1 For the classical helicases (i.e., those which conform to the family name), ATP-binding provides energy to drive the separation of polynucleotide duplexes into single strands. There are also many enzymes which on the basis of structure and motifs appear to be helicases, but which fulfill alternative functions without the necessity for strand separation. These include nucleoprotein remodeling, long-range motion along dsDNA and molecular signaling. The activities and mechanisms of these enzymes, often termed “helicase-like” or “pseudo-helicases,” are not well understood. Recent studies that try to elucidate the full repertoire of helicase activity provide more and more surprises for their numerous roles in genome metabolism.
An example of pseudo-helicase activity is illustrated by the ATP-dependent bacterial Type III restriction endonucleases (REs). For these enzymes, site-specific recognition of a single DNA sequence is insufficient for DNA cleavage. Instead, 2 enzyme complexes that originally bind at separate target sites on the same DNA must interact to produce one double-strand break.Citation2 Since the target sites can be many thousands of base pairs apart, the helicase domains are used to produce “long-range communication” to bring the enzymes close enough for protein–protein interaction and activation of the nuclease domain(s). According to classical helicase mechanisms,Citation1 stepwise motion would require one ATP per base pair moved. Surprisingly, the Type III REs communicate over thousands of base pairs using only tens of ATPs.Citation3 A number of competing models were discussed for the long-range communication of the Type III REs. In addition to dsDNA translocation and 3D DNA looping,Citation4 ATP-independent, random 1D diffusion along the DNA (a.k.a. DNA sliding) was suggested.Citation3
To directly reveal the communication mechanism, we developed a combined magnetic tweezers–total internal reflection fluorescence (TIRF) microscope for the direct visualization of motion by single enzyme complexes. A DNA substrate with 2 type III sites was attached at one end to a glass coverslip and at the other end to a magnetic bead, and was stretched out by the magnetic field so that it was within the evanescent excitation field. The type III RE EcoP15I was labeled with a single fluorescent quantum dot, and the protein trajectory was monitored in real time.Citation5 We were able to observe EcoP15I binding specifically to its site, and, following a short delay, initiation of random 1D motion characteristic of DNA sliding. The diffusion coefficient for this process was one of the largest yet measured, and, given the viscous drag of enzyme and quantum dot, was consistent with uncorrelated motion with respect to the DNA helix. DNA cleavage was directly observed following collision of a sliding enzyme with a second enzyme bound at a distant site.
In combination with ensemble fluorescence measurements, we could reveal that ATP hydrolysis occurred predominantly on the target site and was stringently required to initiate sliding.Citation5 Thus, the helicase domains act as an ATP driven switch. The switching into the sliding state required about 30 ATPs (). A first rapid hydrolysis cycle of 10 ATPs produced a significant conformation change, which we interpret as the formation of a “sliding clamp” structure, and activation of the nuclease domain into a cleavage-competent state. Subsequent hydrolysis of 20 ATPs continued at a slower rate, until the enzyme left the site to start sliding. It is likely that the ATP hydrolysis is used to build up mechanical stress that drives the release of the enzyme from the site. The total number of ATPs required would then reflect uncoupling events, where the helicase transiently disengages from the DNA and another ATP must bind to re-establish the stressed state. Further analysis must, however, illuminate the details of the initiation process.
Figure 1. Models for the ATP-dependent initiation of DNA sliding by type III restriction endonucleasesCitation5 (upper) and the MutS enzymes of mismatch repairCitation6-Citation8 (lower). The type III restriction enzyme EcoP15I comprises 2 Mod subunits (methyltransferase) and 1 Res subunit (helicase in blue, endonuclease in red). MutS enzymes are either homo- or heterodimers. (D = ADP, T = ATP). (1) EcoP15I binds to its target site (5′-CAGCAG-3′), predominantly by a 3D hopping or jumping search pathway, whereas MutS enzymes search for DNA damage by 1D DNA sliding correlated with the DNA helix. (2) Both enzyme systems form a stable complex with their respective target site. The role of ATP is to trigger site release and cause a conformational switch into the activated sliding state necessary for downstream signaling and recruitment of protein co-factors. While EcoP15I requires hydrolysis of ~30 ATPs for this process, MutS proteins require an exchange of 1 or 2 bound ADPs for ATP. While MutS structures indicate how sliding clamps could form, structural information on the type III enzymes is currently lacking. (3) Both enzyme systems enter into a long-lived 1D sliding state. Motion of both classes of enzyme is uncorrelated with respect to the DNA helix, which probably reflects that the downstream interactions are via protein–protein contacts rather than protein–DNA contacts. The MutS diffusion is faster in this step than during the initial mismatch search.
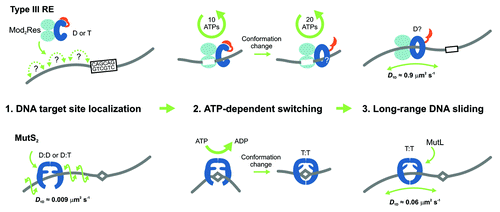
Our study revealed a new functionality for helicases, namely as a molecular switch, which, in this case, initiates long-lived DNA sliding. We expect that more pseudo-helicases will be found that act as molecular switches in other processes, where latent activities need to be triggered following a recognition event. A similar sliding-based communication mechanism has now been demonstrated for the mismatch repair protein MutS, where nucleotide exchange (rather than hydrolysis) in its ABC transporter ATPase domains triggers sliding on DNA after mismatch recognition ().Citation6-Citation8 The sliding of MutS and also of its eukaryotic counterparts is used to search for strand discrimination signals on DNA that identify the damaged strand. Thus, ATP-triggered signaling through sliding is becoming a more widespread phenomenon. The difference in ATP consumption between type III REs and MutS may be due to the different ATPase classes involved. We suggest that the necessity for an ATP-triggered step to initiate sliding reflects the need for stringent control of downstream processes that in both cases results in DNA cleavage. This can be compared with the purchase of a ticket with which one is allowed to enter into the next phase. Such nucleotide-based licensing seems to provide an overall benefit to biological processes. For example, nucleotide hydrolysis is also used during translation for proofreading the binding of the correct tRNA.
References
- Singleton MR, et al. Annu Rev Biochem 2007; 76:23 - 50; http://dx.doi.org/10.1146/annurev.biochem.76.052305.115300; PMID: 17506634
- Meisel A, et al. Nature 1992; 355:467 - 9; http://dx.doi.org/10.1038/355467a0; PMID: 1734285
- Ramanathan SP, et al. Proc Natl Acad Sci U S A 2009; 106:1748 - 53; http://dx.doi.org/10.1073/pnas.0807193106; PMID: 19181848
- Dryden DT, et al. Nucleic Acids Res 2011; 39:4525 - 31; http://dx.doi.org/10.1093/nar/gkq1285; PMID: 21310716
- Schwarz FW, et al. Science 2013; 340:353 - 6; http://dx.doi.org/10.1126/science.1231122; PMID: 23599494
- Qiu R, et al. EMBO J 2012; 31:2528 - 40; http://dx.doi.org/10.1038/emboj.2012.95; PMID: 22505031
- Gorman J, et al. Proc Natl Acad Sci U S A 2012; 109:E3074 - 83; http://dx.doi.org/10.1073/pnas.1211364109; PMID: 23012240
- Cho WK, et al. Structure 2012; 20:1264 - 74; http://dx.doi.org/10.1016/j.str.2012.04.017; PMID: 22682745