Abstract
During mouse oocyte maturation histones are deacetylated, and inhibiting this deacetylation leads to abnormal chromosome segregation and aneuploidy. RBBP7 is a component of several different complexes that contain histone deacetylases, and therefore could be implicated in histone deacetylation. We find that Rbbp7 is a dormant maternal mRNA that is recruited for translation during oocyte maturation to regulate the histone deacetylation. Importantly, we show that the maturation-associated decrease of histone acetylation is required for localization and function of the chromosomal passenger complex (CPC) during oocyte meiotic maturation. This finding can explain the phenotypes of oocytes where Rbbp7 is depleted by an siRNA/morpholino cocktail including severe chromosome misalignment, improper kinetochore–microtubule attachments, impaired SAC function, cytokinesis defects, and increased incidence of aneuploidy at metaphase II (Met II). These results implicate RBBP7 as a novel regulator of histone deacetylation during oocyte maturation and provide evidence that such deacetylation is required for proper chromosome segregation by regulating localized CPC function.
Introduction
In mammals, oocyte maturation involves coupling the meiotic cell cycle with key developmental events. One such event is global histone deacetylation, which is critical for normal chromosome condensation and alignment on the spindle and, ultimately, for generating eggs with proper chromosome numbers. Acetylation of histones H3 and H4 is controlled by histone acetyl transferases (HATs) and histone deacetylases (HDACs). Although HATs are active in prophase I-arrested oocytes, HATs are inactive and cannot acetylate histone H3 and H4 during meiosis I (MI).Citation1 Instead, HDAC activity predominates and deacetylates histones during MI.Citation1 Thus, HDACs are presumably the central players controlling the acetylation status of histones during oocyte maturation. Our previous studies indicate that HDAC2 is the central HDAC functioning in mouse oocytes.Citation2 Whether HDAC activity is regulated during maturation, and if so, what the basis for that regulation is, is not known.
Another developmental event that occurs during oocyte maturation is the recruitment of RNAs stored within the egg for translation, which ensures that the egg contains enough proteins to support meiosis, egg development, fertilization, and early embryonic events. Retinoblastoma binding protein P46 (RBBP7), also known as RBAP46, is one of many of these maternally recruited messages identified through microarray experiments.Citation3 RBBP7 is a ubiquitously expressed nuclear protein that belongs to the WD-40 protein family and is an integral subunit in HDAC complexes, ATP-dependent nucleosome-remodeling factor (NURF) complexes, and EZH2/EED methylase complexes. Within HDAC complexes such as SIN3ACitation4,Citation5 and NuRD (nucleosome remodeling and HDAC),Citation6,Citation7 RBBP7 binds directly to helix 1 of histone H4. RBBP7 also binds histone H2A in yeast and mammalian cells, albeit to a lesser extent than H4.Citation8 One function implicated within the NURF complexes is transcriptional regulation.Citation9 During preimplantation embryonic development RBBP7 is found within the EZH2/EED complexes that methylate either histone H3K27 or histone H1K26 to promote epigenetic silencing of regulatory genes.Citation10,Citation11 The significance of RBBP7 maternal recruitment for oocyte maturation is not known.
All HDACs lack a DNA binding domain and execute their function by being a component of repressor complexes.Citation12 Given that RBBP7 can regulate HDAC activity by providing HDACs the ability to bind to chromatin, we explored a possible role for RBBP7 in the maturation-associated histone deacetylation during oocyte maturation. Using an siRNA/morpholino knockdown approach, we found that RBBP7 depletion inhibits the maturation-associated decrease in acetylation of histone H3 and H4, and that the maturation-associated increase in RBBP7 protein is critical for generating euploid eggs by regulating formation of stable kinetochore–microtubule connections. Importantly, we find that chromosomal passenger complex (CPC) localization and activity are ablated in RBBP7-depleted oocytes. Meiotic maturation is partially rescued when Aurora kinase C (AURKC), the catalytic component of the CPC, is overexpressed in knockdown oocytes, indicating that the phenotypic observations are, at least in part, due to altered localized AURKC function. These data reveal new insights connecting regulation of AURKC-CPC function and its role in controlling chromosome segregation during MI with histone deacetylation.
Results
Rbbp7 is a maternally recruited message
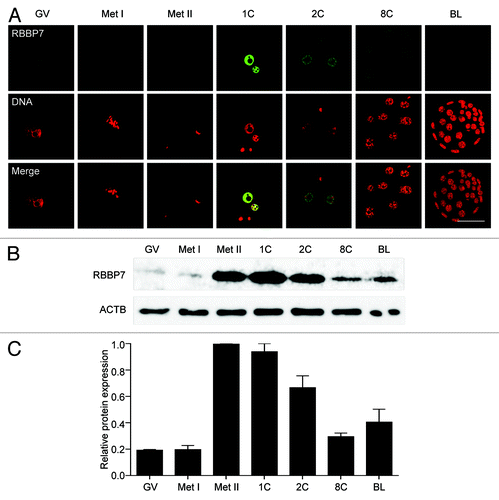
In a microarray study of mouse oocytes, Rbbp7 is a message that is likely recruited for translation during meiotic maturation.Citation3 To confirm this result at the protein level, we performed immunocytochemistry to detect RBBP7 protein during oocyte maturation and preimplantation embryogenesis. RBBP7 protein localized to the nucleus (germinal vesicle, GV), as anticipated, but displayed a maturation-associated increase in protein abundance that was clearly evident in 1-cell embryos (). Upon meiotic maturation, local protein concentration can be diluted 20–30-fold when the nuclear envelope breaks down. When the confocal laser power was increased to detect the signal in metaphase I (Met I) and metaphase II (Met II) eggs, although the bulk of the signal was cytoplasmic, some RBBP7 localized with the spindle (Fig. S1). Immunoblotting confirmed the maturation-associated increase in RBBP7 and further showed that the increase occurred between Met I and Met II (). Consistent with the quantitative RT-PCR profile of Rbbp7 transcript (Fig. S2) and immunocytochemistry results, immunoblotting revealed a decrease in protein commencing at the 1-cell stage followed later with an increase upon zygotic expression at the blastocyst stage. The maturation-associated increase in RBBP7 indicates that it is another member of a growing list of maternal mRNAs that is recruited during oocyte maturation.Citation13
Figure 1. Temporal and spatial pattern of RBBP7 expression during oocyte maturation and preimplantation embryo development. (A) Immunocytochemical analysis of RBBP7 expression. All samples were processed for immunocytochemistry together, and all images were taken at the same laser power. The experiment was conducted 3 times, and at least 20 oocytes/embryos were analyzed for each sample. Shown are representative examples. The scale bar represents 50 μm. (B) Immunoblot analysis of RBBP7 expression. Forty oocytes/embryos were loaded per lane, and the experiment was conducted twice, and β-actin (ACTB) was used as a loading control. Shown is a representative example. (C) Corresponding histogram of relative immunoblot analysis and the data are expressed as mean ± SEM. GV, germinal vesicle; Met I, metaphase I; Met II, metaphase II; 1C, 1-cell embryo; 2C, 2-cell embryo; 8C, 8-cell embryo; BL, blastocyst.
siRNA and morpholino-mediated knockdown of RBBP7 perturbs histone deacetylation
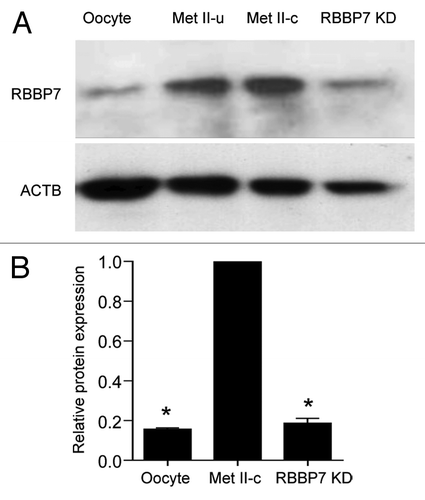
To address the role of RBBP7 during oocyte maturation, full-grown, nucleus-intact oocytes were injected with a mixture of siRNA and morpholino oligonucleotides. This dual approach was necessary to effectively deplete the RNA stores of Rbbp7 and to block translational recruitment of the remaining messages. The maturation-associated increase in RBBP7 protein was almost totally inhibited by this approach ().
Figure 2. siRNA and morpholino mediated knockdown of RBBP7. (A) Immunoblot analysis of RBBP7 expression. Full-grown oocytes were injected with combination of siRNA and morpholino. After 18 h, 40 eggs were used for immunoblot analysis and β-actin (ACTB) was used as a loading control. The experiment was performed 6×. Shown is a representative example. Met II-u is 40, uninjected eggs; Met II-c is 40 control injected eggs. (B) Corresponding histogram of relative immunoblot analysis of (A). One-way ANOVA was used to analyze the data. The data are expressed as mean ± SEM. Values with an asterisk vary significantly, P < 0.05.
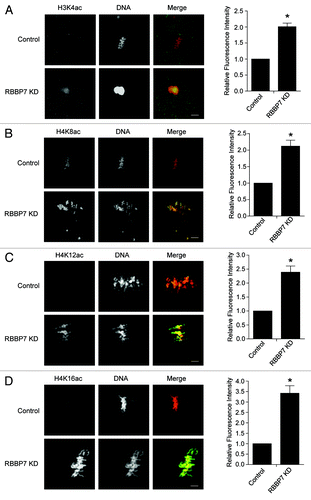
Next, to assess a role for RBBP7 in regulating histone deacetylation, we assayed different histone acetylation marks in knockdown (KD) oocytes during maturation using immunocytochemistry. Results of these experiments indicated that deacetylation of H3K4, H4K8, H4K12, and H4K16 was significantly inhibited following RBBP7 KD (). Acetylation of these lysines increased 2- to 3-fold compared with controls. There was no apparent effect on deacetylation of H3K9, H3K14, or H4K5 (Fig. S3). These results suggest that RBBP7 regulates HDAC activity during oocyte maturation.
Figure 3. Effect of RBBP7 knockdown on histone acetylation status in MII oocytes. Immunocytochemical detection of H3K4ac (A), H4K8ac (B), H4K12ac (C), and H4K16ac (D) in RBBP7 KD Met II eggs. The experiments were performed 3×, and at least 20 oocytes were analyzed for each sample. The scale bars represent 10 μm. Shown are representative examples. The graphs on the right of the images are average quantifications of the pixel intensity of the indicated histone marks to the left. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with an asterisk vary significantly, P < 0.05.
RBBP7 depletion affects meiotic progression and chromosome segregation
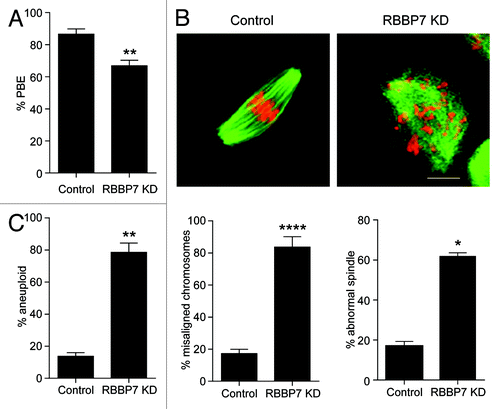
To determine the biological significance of RBBP7-dependent histone deacetylation during oocyte maturation, we injected oocytes with the siRNA/morpholino cocktail and matured the oocytes to Met II. Compared with injected controls, there was a significant but modest decrease in polar body emission (). When the Met II eggs were fixed and processed for immunocytochemistry to evaluate their spindles and DNA morphology, we observed surprisingly striking defects. Although many RBBP7-depleted oocytes extruded polar bodies, there was a significant number that contained severely misaligned chromosomes and abnormal spindles (). Therefore, as anticipated, the incidence of aneuploidy in Met II eggs was significantly higher following RBBP7 knockdown compared with controls (). Taken together, these data suggest that events occurring during MI are perturbed in RBBP7-depleted oocytes.
Figure 4. Effect of RBBP7 knockdown on meiotic progression. (A–C) Full-grown oocytes were injected with combination of siRNA and morpholino followed by in vitro maturation for 18 h and scored for first polar body extrusion (A) followed by fixation and staining with an anti-β-tubulin antibody (green) to detect spindles and TOPRO-3 to detect DNA (red) using confocal microscopy (B). The confocal images were analyzed for the presence of misaligned chromosomes and to detect abnormal spindle morphology. (C) MII eggs were treated with monastrol for 2 h prior to fixation and detection of kinetochores with Crest anti-serum and DNA with TOPRO-3. Eggs with greater or less than 40 kinetochores were considered aneuploid. The experiment was performed 3×, and at least 30 oocytes were examined for each sample. The scale bar represents 100 μm. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, **P < 0.01, ****P < 0.0001.
To visualize how the Met II spindle and chromosome defects arose, we used time-lapse confocal imaging. To mark spindles, we injected oocytes with Aurka-Gfp cRNA that localizes to spindle poles,Citation14 and, to monitor chromosome movement, we co-injected cRNA encoding H2b-mCherry (Video S1). Depleting RBBP7 resulted in multiple phenotypes. We observed cytokinesis defects that we could not detect in our fixed images. The most common cytokinesis phenotype was a ruffling in the appearance of the cell membrane during anaphase, prior to polar body emission (; Video S2). Moreover, we observed cytokinesis failure in which the oocytes did attempt extrusion of a polar body, but this was followed by its retraction (; Video S3). To our surprise, in these instances, the chromosomes collapsed after cytokinesis (~21% of oocytes). Likely underlying the collapse was a failure to form a stable spindle, because there was a lack of detectable AURKA-GFP on anything resembling a spindle pole in these oocytes. Consistent with the Met II misalignment phenotype, we observed a failure of chromosomes to align on the Met I plate (). Therefore, chromosome alignment and cytokinesis are compromised in RBBP7-depleted oocytes.
Figure 5. Time-lapse confocal observations of meiosis following RBBP7 knockdown. (A) Full-grown oocytes were injected with combination of siRNA and morpholino, and cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (green) followed by in vitro maturation for 18 h. Bright field and fluorescence photographs were acquired every 20 min. Examples of observed abnormal cytokinesis are noted in Telo I. The scale bar represents 50 μm. (B) Quantification of abnormal cytokinesis defects in (A). GV, germinal vesicle intact oocyte; Pro-Met I, prophase I-metaphase I); Ana I, anaphase I; Telo I, telophase I; Met II, metaphase II. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, ***P < 0.001.
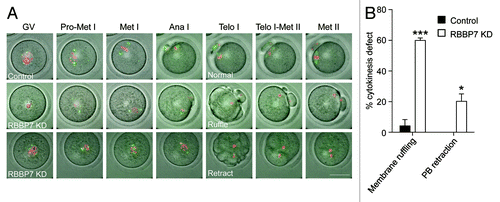
Figure 6. Effect of RBBP7 knockdown on spindle formation, chromosome alignment, and K-MT attachment. (A) Microtubules (green) and DNA (red) from the indicated treatment group were stained at Met I and examined for spindle formation and chromosome alignment by immunocytochemistry. The experiment was conducted 3×, and at least 20 oocytes were analyzed for each sample. The scale bar represents 10 μm. Shown are representative examples. (B) Corresponding quantification of (A). Student t test was used to analyze the data. (C) Representative confocal images from the cold-stable microtubule assay from the indicated treatment groups. Kinetochores were detected with Crest anti-sera (red), microtubules with an anti-β-tubulin antibody (green) and DNA with DAPI (blue). The insets highlight normal (control panel), abnormal (KD panel 1), or unattached (KD panel 2) kinetochores. The scale represents 10 μm. (D) Quantification of the types of attachments observed. At least 750 kinetochores were analyzed from 2 different experiments. The data are expressed as mean ± SEM; One-way ANOVA was used to analyze the data. Values with asterisks vary significantly, ***P < 0.001, ****P < 0.0001.
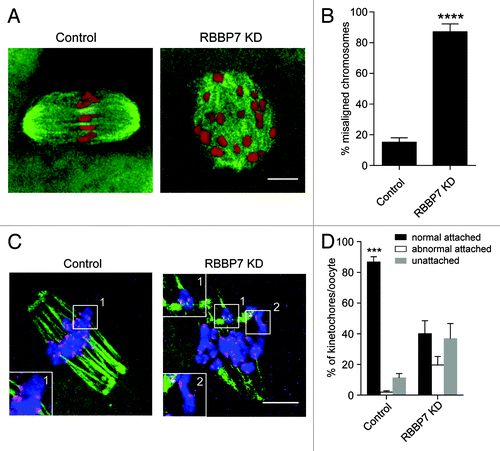
The high incidence of chromosome misalignment could reflect reduced ability of the kinetochores to form stable microtubule attachments; such microtubules are cold-stable, whereas microtubules that do not form stable attachments with the kinetochore are cold-labile.Citation15 In somatic cells, histone hyperacetylation can interfere with kinetochore assembly,Citation16 and in budding yeast, hypoacetylation of H4K16 is critical to maintain kinetochore function.Citation17 Moreover, in oocytes from Hdac2−/− mice where H4K16 is hyperacetylated microtubules fail to make stable attachments to kinetochores.Citation2 Accordingly, we analyzed whether targeting RBBP7 resulted in fewer cold-stable microtubule–kinetochore attachments. In controls most kinetochores (detected by CREST anti-serum) had clearly associated microtubules (). In contrast, many kinetochores in the experimental group either did not have attached microtubules or contained improper attachments, although kinetochores themselves appeared structurally intact, as determined by HEC1 staining (Fig. S4). This finding is consistent with the high incidence of aneuploidy observed following targeting RBBP7 ().
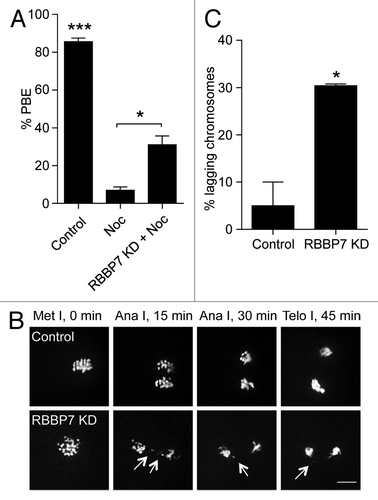
The observation that there is a significant number of unattached kinetochores, but that cytokinesis still proceeds suggests the surveillance mechanism (spindle assembly checkpoint, SAC) that prevents anaphase onset until proper attachments are made is not functioning. To assess the strength of the SAC, we incubated oocytes in a low dose of nocodazole, a microtubule-depolymerizing agent that activates the SAC, and assessed polar body extrusion. If the SAC is functional, the oocytes should maintain arrest at Met I and not extrude a polar body. Consistent with our hypothesis, ~35% of RBBP7 KD oocytes extruded polar bodies (), indicating a weakened SAC. When chromosome segregation occurs without proper K–MT attachments, these chromosomes can often be observed segregating abnormally. When we looked at RBBP7-KD oocytes live, we observed a higher frequency of lagging chromosomes during Ana I-Telo I compared with controls (). Taken together, multiple events that occur during MI are perturbed in RBBP7-depleted oocytes. These perturbations likely contribute to increased levels of aneuploid Met II eggs ().
Figure 7. Effect of RBBP7 knockdown on the spindle assembly checkpoint. (A) Full-grown oocytes were injected with combination of siRNA and morpholino. Control-injected oocytes were incubated in maturation medium plus/minus 400 nM nocadozole (Noc). After 16 h of in vitro maturation, extrusion of the polar body was confirmed using light microscopy. The data are expressed as mean ± SEM; One-way ANOVA was used to analyze the data. (B) Representative examples of live imaging of oocytes from the indicated treatment groups co-injected with H2b-mCherry cRNA. The arrows point to lagging chromosomes. The scale bar represents 10 μm. (C) Quantification of (B). The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, ***P < 0.001.
CPC localization is perturbed in RBBP7 knockdown oocytes
In mitosis, Aurora kinase B (AURKB), the catalytic subunit of the chromosomal passenger complex (CPC), regulates chromosome congression, the stability of microtubule–kinetochore interactions, the spindle assembly checkpoint, and cytokinesis.Citation18-Citation21 The CPC is therefore a critical regulator of chromosome segregation and genome integrity.Citation22-Citation24 To execute this function, the CPC localizes to centromeres. AURKB has specificity for hypoacetylated histone H3, and it is thought that the chromatin remodeling that occurs during G2 may set up a chromosome state that favors downstream kinetochore assembly.Citation25,Citation26 Oocytes contain an AURKB homolog called AURKC that appears to have similar functions as AURKB in the CPC.Citation14,Citation27-Citation29 The CPC localizes to centromeres and inter-chromatid axes, where it promotes proper chromosome segregation during MI and to kinetochores at Met II.Citation14,Citation29,Citation30 The defects that we observed in RBBP7-depleted oocytes are strikingly similar to defects in oocytes, where components of CPC are perturbed.Citation14,Citation30-Citation32 We therefore first assessed the subcellular localization and activity of the CPC in the RBBP7 KD oocytes at Met I via immunocytochemistry. AURKC localized to the interchromatid axes in control-injected oocytes, as previously reported.Citation14,Citation29,Citation30 In contrast, in RBBP7 KD oocytes, such localization of AURKC was not detected (). AURKC kinetochore localization was also not detected in KD oocytes that were matured to Met II (Fig. S5A), whereas AURKC-GFP primarily localized to the meiotic spindle when expressed in the KD oocytes (). We did note that a weak GFP signal could be detected at some kinetochores and may reflect high levels of overexpression. These data indicate that the localization of the catalytic subunit of the CPC is altered in RBBP7-depleted oocytes.
Figure 8. Effect of RBBP7 knockdown on the CPC during MI. Immunocytochemical detection of AURKC (A and B), Survivin (C), and pINCENP (D) (green in merge) in control and RBBP7 KD Met I oocytes. DNA was detected with DAPI (red in merge). In (B), 100 ng/μl AURKC-GFP (green in merge) was co-injected with the siRNA/morpholino cocktail. To the right of the image panels are quantifications of the intensity levels (A and D) or the percent of oocytes where normal chromosome localization was observed (B and C). The experiments were performed 2×, and at least 15 oocytes were analyzed for each sample. The scale bar represents 10 μm. Shown are representative examples. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, * P < 0.05, **P < 0.01.
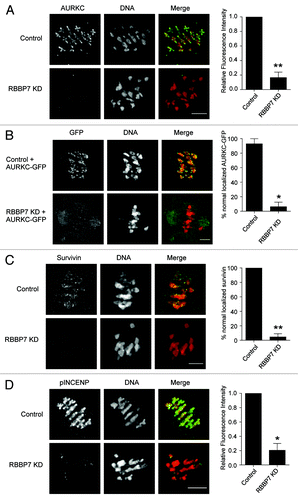
Survivin is another component of the CPC. Consistent with delocalized AURKC, survivin failed to localize in RBBP7-depleted oocytes (). Within the CPC, AURKC phosphorylates inner-centromere protein (INCENP).Citation33,Citation34 Using an antibody that specifically recognizes the phosphorylated AURKB/C residues in INCENP (pINCENP), we found that in contrast to control oocytes, pINCENP levels in RBBP7 KD oocytes were greatly diminished (; Fig. S5B). These data indicate that CPC activity is reduced. To confirm that this effect on the CPC was due to a change in histone acetylation, and not because of a direct recruitment of the CPC by RBBP7, we treated oocytes with trichostatin A (TSA), a class I and II HDAC inhibitor. Compared with controls, we found diminished chromosome-localized AURKC and pINCENP signals (Fig. S5C and D). Therefore, hyperacetylation of histones through RBBP7 depletion alters the ability of the CPC to localize to chromosomes.
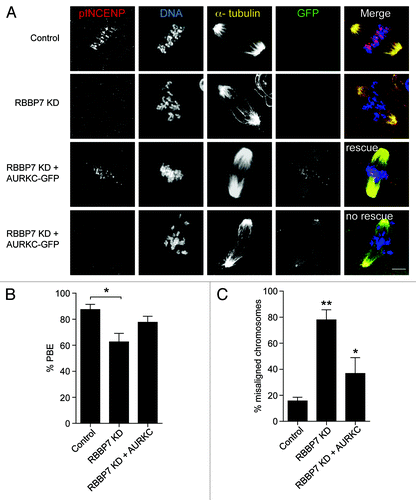
Finally, to confirm that lack of localized CPC function causes the observed phenotypes, we overexpressed AURKC-GFP (200 ng/μl) in RBBP7-depleted oocytes. In ~69% of the oocytes, the exogenous AURKC-GFP relocalized to chromosomes and rescued the misalignment and polar body extrusion phenotypes at Met II (). In the remaining oocytes in which AURKC-GFP failed to localize, we did not detect a pINCENP signal, and the phenotypic rescue failed to occur. Taken together, these data suggest histone hyperacetylation perturbs localized AURKC–CPC function.
Figure 9. Overexpression of AURKC rescues the defects observed in RBBP7 knockdown oocytes. Full-grown oocytes were injected with the indicated materials and matured in vitro for 18 h, prior to fixation and detection of phosphorylated INCENP (pINCENP; red), the spindle (α-tubulin; yellow), and DNA (DAPI; blue). (A) Representative confocal images of each treatment group. In the “rescue” group, AURKC-GFP signal was found at kinetochores (green), whereas in the “no rescue” group, AURKC-GFP signal was at the spindle poles. Scale bar represents 10 μm. (B and C) Quantification of the percentage of oocytes in (A) that extruded a polar body (PBE), and had chromosome misalignment, respectively. The experiments were performed 3×, and at least 15 oocytes were analyzed for each sample. The data are expressed as mean ± SEM; One-way ANOVA was used to analyze the data. Values with asterisks vary significantly, * P < 0.05, **P < 0.01.
Discussion
During oocyte maturation, histone deacetylation is essential for accurate chromosome segregation that is required to generate healthy, developmentally competent eggs.Citation2 But it was not known how HDACs are regulated during oocyte maturation. We report that the histone chaperone RBBP7Citation35 regulates histone deacetylation during oocyte meiotic maturation (minimally at H3K4, H4K8, H4K12, and H4K16). RBBP7 is presumably regulating the activity of HDACs present in complexes, because in mitosis, RBBP7 co-immunoprecipitates with HDAC complexes.Citation6,Citation36,Citation37 In this study, we investigated the roles of RBBP7 in mouse oocyte meiosis. Perturbing the maturation-associated decrease in histone acetylation during oocyte maturation results in loss of localized CPC function, a situation that leads to the observed inaccurate chromosome segregation. This is the first report to provide mechanistic insights concerning how perturbing HDAC activity through RBBP7 during oocyte maturation compromises egg quality.
A higher signal of acetylated H4K8 and H4K12 is detected in MII eggs from older mice than from young mice and is correlated with high levels of aneuploidy.Citation38 Deletion of HDAC2 during oocyte maturation leads to hyperacetylation of H4K16, with subsequent aneuploidy in Met II eggs.Citation2 In our study, not only are H4K8, H4K12, and H4K16 hyperacetylated, but so is histone H3K4 (). These findings suggest that RBBP7 regulates at least one other HDAC complex, in addition to likely regulating HDAC2-containing complexes.Citation6,Citation36 Identifying these HDACs will likely give us insight in age-related increases in aneuploidy that appear to be associated with histone acetylation changes.
The phenotype of RBBP7 KD is consistent with the phenotype that occurs when histones H3 and H4 are hyperacetylated during mouse oocyte maturation through other perturbations. Inadequate histone deacetylation of H4K12 during oocyte meiosis is associated with chromosome misalignment and aneuploidy in mice.Citation38 Moreover, global inhibition of all HDAC activity by trichostatin A treatment during mouse oocyte maturation results in severe chromosome misalignment (>75%), lagging chromosomes, abnormal chromatid separation during cytokinesis, abnormal chromatid separation during cytokinesis.Citation39 Treatment of tumors with OSU-HDAC-44, an HDAC inhibitor, reduces both AURKB and Survivin levels and causes cytokinesis defects.Citation40 These observations are supported by the defective chromosome segregation and kinetochore function in Hdac2−/− oocytes.Citation2 Although it is known that hyperacetylation of histones interferes with kinetochore assembly and function in mitosis,Citation16,Citation17 the mechanism by which the hyperacetylation perturbs kinetochore function during oocyte maturation is still unclear.
In mitosis, the CPC is essential for chromosome segregation by regulating K–MT attachments, activating the spindle assembly checkpoint and regulating cytokinesis.Citation18-Citation21 It is important to note that although AURKB is most commonly thought to destabilize improper K-MTs, at least 2 recent reports demonstrate that loss of AURKB activity results in loss of K–MT interactions,Citation22,Citation41 similar to our observations. One way the CPC catalytic subunit, AURKB, regulates K-MT interaction is by recruiting motor proteins dynein and CENP-E to kinetochores.Citation22 In germ cells, AURKC replaces AURKB in the CPC and is essential for faithful chromosome segregation during mouse oocyte maturation.Citation14,Citation29,Citation30,Citation32 To our knowledge, there are no available data regarding the possible relationship between histone deacetylation and Aurora kinase function or AURKC in meiosis. Importantly, we find complete loss of localized CPC activity () when histones are hyperacetylated during oocyte meiosis. These results clearly show that HDAC activity is a prerequisite for regulating AURKC–CPC localization to ensure normal chromosome segregation and cytokinesis and are consistent with data showing that phosphorylation of H3S10, another readout of AURK activity, is reduced in oocytes cultured in TSA.Citation39 Furthermore, these findings support previous observations that AURKB-CPC has higher specificity for hypoacetylated H3 during mitosis.Citation26 An alternative model could be that AURKC is directly acetylated, thereby perturbing its activity and localization.Citation25,Citation26 However, it is not known if AURKC is regulated by acetylation during meiosis. The effect of histone hyperacetylation on localized AURKC function readily explains the RBBP7 phenotypes we observe, and also unveils another regulator of the CPC kinase in mouse oocyte meiosis. To our knowledge, this is the first report to show a connection between histone acetylation and CPC function during meiotic maturation. Therefore, further understanding of how histone deacetylation regulates the meiotic CPC will be critical to determining how MI is regulated in female meiosis.
Materials and Methods
In vitro cRNA synthesis
Generation of Aurka-Gfp, Aurkc-Gfp, and H2b-mCherry were described previously.Citation14,Citation29 DNA linearization of all Gfp- and mCherry-containing constructs was performed using Nde I (New England BioLabs). After DNA linearization, the digests were purified (Qiagen, QIAquick PCR Purification), and in vitro transcription was performed using an mMessage mMachine T7 kit (Ambion) according to the manufacturer’s instructions. Finally, the cRNA was purified using an RNAEasy kit (Qiagen).
Oocyte collection, microinjection, treatment, and in vitro maturation
Full-grown GV-intact oocytes freed of attached cumulus cells were obtained from eCG primed 6-wk-old female mice as previously described.Citation42 The collection and injection medium for oocytes was bicarbonate-free minimal essential medium (Earle salt) containing, 25 mM Hepes, pH 7.3, 3 mg/ml polyvinylpyrollidone (MEM/PVP), and 2.5 μM milrinone to prevent meiotic resumption.Citation43
Oocytes were microinjected with 10 pl of a combination of siRNA (25 μM) and morpholino (1 mM) for Rbbp7. siRNAs to target Rbbp7 (5′-UUUCAGAUUA CGCAGGUCCC A-3′) (Ambion, Inc) were diluted with MilliQ water to a final concentration of 100 μM and stored at −80 °C. The morpholino oligonucleotide spanning the start codon of Rbbp7 transcript (5-CTTCAAACAT CTCTTTACTC GCCAT-3′) was purchased from Gene Tools LLC (Philomath). Control oocytes were injected with a combination of a siRNA (Luciferase GL2 Duplex; D-001100-01-05, Thermo Fisher Scientific) and morpholino (5′-CCTCTTACCT CAGTTACAAT TTATA-3′). Following microinjection, the oocytes were cultured in Chatot, Ziomek, and Bavister (CZB)Citation44 medium containing 2.5 μM milrinone under 5% CO2 in air at 37 °C for 1 h. The oocytes were then matured in vitro in milrinone-free CZB medium for either 8 h or 18 h in 5% CO2 in air at 37 °C.
Histone deacetylase inhibitor Trichostatin A (TSA, Sigma T8552) was added to the maturation medium at concentration of 1 μM under the same culture condition for 8 h.
All animal experiments were approved by the institutional animal use and care committee and were consistent with National Institute of Health (NIH) guidelines.
Real-time PCR
Fifty oocytes or embryos at the indicated developmental stage were isolated and frozen at −80 °C prior to processing. After thawing on ice, 2 ng of Gfp cRNA was added to each sample. Total RNA was purified using the Picopure RNA isolation kit (Arcturus) according to the manufacturer’s protocol. Reverse transcription, primed with random hexamers, was performed using superscript II reverse transcriptase (Invitrogen) following the manufacturer’s instructions. TaqMan probes specific for Rbbp7 transcripts (Applied Biosystem) were used, and the comparative Ct method was used to determine differences in expression levels between stages. Data were acquired using an ABI prism 7000 (Applied Biosystem).
Immunoblotting
Oocytes or embryos were lysed in 1% SDS, 1% β-mercaptoethanol, 20% glycerol, and 50 mM Tris–HCl (pH 6.8), and denatured at 95 °C for 5 min. Proteins separated by electrophoresis in 10% gradient SDS polyacrylamide precast gel. Stained proteins of known molecular mass (range: 14–200 kDa) were run simultaneously as standards. The electrophoretically separated polypeptides were transferred to nitrocellulose membranes, which were then blocked by incubation in 2% blocking (ECL blocking; Amersham) solution in TBS-T (Tris-buffered saline with 0.1% Tween 20) for 1 h. The membranes were then incubated with primary antibodies at 4 °C overnight. After washing with TBS-T 5×, the membranes were incubated with a secondary antibody labeled with horseradish peroxidase for 1 h followed with washing with TBS-T 5×. The signal was detected using the ECL Advance western blotting detection reagents (Amersham) following the manufacturer’s protocol.
Immunocytochemistry
For analysis of cold-stable microtubules, oocytes were incubated for 10 min on ice in MEM/PVP containing 20 mM Hepes, pH 7.3, and then fixed for 25 min on a warm plate with 3.7% formaldehyde in 100 mM Pipes, pH 6.8, containing 10 mM EGTA, 1 mM MgCl2, and 0.2% Triton X-100.Citation45 For all other experiments, oocytes or embryos were fixed in 2.5% paraformaldehyde in PBS for 20 min at room temperature. After fixation, the cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min and transferred to blocking buffer (PBS + 0.3% BSA+ 0.01% Tween-20) for 15 min. Immunostaining was performed by incubating the fixed cells with the primary antibody overnight at 4 °C, followed by secondary antibodies conjugated with Alexa 488 for 60 min; omission of the primary antibody served as negative control. DNA was stained and mounted with TO-PRO-3 (Life Technologies, Grand Island, T3605; 1:500) diluted in VectaShield (Vector Laboratories) under a coverslip with gentle compression. Fluorescence was detected on a Leica TCS SP or a Zeiss 510 meta (; Fig. S3) laser-scanning confocal microscopes.
For RBBP7 immunostaining, all samples, i.e., oocytes, eggs and embryos, were processed at the same time. The laser power was adjusted to a level where signal intensity was just below saturation for the developmental stage that displayed the highest intensity, and all images were then scanned at that laser power. The intensity of fluorescence was quantified with NIH imageJ software.
In situ chromosome counting
Monastrol treatment, immunocytochemical detection of kinetochores, and chromosome counting were performed as previously described.Citation46,Citation47 Briefly, eggs were cultured for 1 h in CZB containing 100 μM monastrol (Sigma) to disperse the chromosomes by collapsing the bipolar spindle to a monoploar spindle. Eggs were fixed in freshly prepared 2.5% paraformaldehyde and stained for kinetochores (CREST antibody) and DNA (Sytox Green; Invitrogen, A11012; 1: 5000). Images were collected at 0.4-μm intervals to span the entire region of the MII spindle (16–20 μm total) using a Leica DM6000 microscope with 100× 1.4 NA oil immersion objective. To obtain a chromosome count for each egg, serial confocal sections were analyzed to determine the total number of kinetochores and calculated using NIH ImageJ software.
Live cell imaging
Oocytes (previously microinjected with cRNAs encoding an AURKA-GFP fusion protein and a histone H2B-mCherry fusion protein) were transferred into separate drops of CZB medium covered with mineral oil in a FluoroDish (World Precision Instruments). DIC, GFP and mCherry image acquisition was started at the GV stage using a Leica DM6000 microscope with a 63× 1.25 NA oil immersion objective and a charge-coupled device camera (Orca-AG, Hamamatsu Photonics) controlled by Metamorph Software. The microscope stage was heated to 37 °C, and 5% CO2 was maintained using a microenvironment chamber (PeCon) and an airstream incubator (ASI 400, Nevtek). Images of individual cells were acquired every 20 min and processed using NIH ImageJ software.
Antibodies
The following antibodies were used in immunofluorescence (IF) and/or western blotting (WB): anti-RBBP7 (ab3535; Abcam; WB, 1:10 000; IF, 1:500), anti-β-actin (ab20272; Abcam; WB, 1:10 000), anti-β-tubulin (3623, Danvers, cell signaling; IF, 1:75), anti-α-tubulin-Alexa Fluor 488 conjugate (Life Technologies #322588; 1:100), CREST autoimmune serum (Immunovision; IF, 1:40), anti-AURKC (A400-023A- BL1217; Bethyl; IF, 1:30), anti-survivin (Cell Signaling Technology #2808S; 1:500), phospho-specific S893/S894 INCENP (kind gift of Michael Lampson, UPenn;Citation33 IF, 1:1000), anti-HEC1 (kind gift of Robert Benezra, Memorial Sloan-Kettering Cancer Center;Citation48 IF, 1:500), anti-acetyl-histone H3 (Lys4) (39382; active motif; IF, 1:200), and anti-acetyl-histone H3 (Lys14) (A-4023; Epigentek; IF, 1:200). Anti-acetyl-histone H3 (Lys9) (04-1003), anti-acetyl-histone H4 (Lys5) (06-759), anti-acetyl-histone H4 (Lys8) (06-760), anti-acetyl-histone H4 (Lys12) (06-761), and anti-acetyl-histone H4 (Lys16) (06-762) were all from Millipore and used in IF at 1:300.
Statistical analysis
Student t test and 1-way ANOVA was used to evaluate the differences between groups. The differences of P < 0.05 were considered significant.
| Abbreviations: | ||
| Ana I | = | anaphase I |
| CPC | = | chromosomal passenger complex |
| GV | = | germinal vesicle |
| HAT | = | histone acetyltransferase |
| HDAC | = | histone deacetylase |
| MI | = | meiosis I |
| Met I | = | metaphase I |
| Met II | = | metaphase II |
| Telo I | = | telophase I |
| TSA | = | trichostatin A |
Additional material
Download Zip (9.3 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by grants from the NIH (HD022681 to RMS and HD61657 to KS) and RU start-up funds to KS. AZB was supported by a fellowship funded from Science and Technology Development Fund (US-Egypt Joint Science and Technology Board) and RU start-up funding. The authors thank Michael Lampson for the pINCENP antibody, Robert Benezra for the HEC1 antibody, and Pengpeng Ma, Richard Jimenez, Sergey Medvedev, Jun Ma, and Olga Davydenko for their valuable discussions and help.
References
- Kim JM, Liu H, Tazaki M, Nagata M, Aoki F. Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol 2003; 162:37 - 46; http://dx.doi.org/10.1083/jcb.200303047; PMID: 12835313
- Ma P, Schultz RM. Histone deacetylase 2 (HDAC2) regulates chromosome segregation and kinetochore function via H4K16 deacetylation during oocyte maturation in mouse. PLoS Genet 2013; 9:e1003377; http://dx.doi.org/10.1371/journal.pgen.1003377; PMID: 23516383
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol 2004; 272:483 - 96; http://dx.doi.org/10.1016/j.ydbio.2004.05.018; PMID: 15282163
- Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol 1999; 19:5847 - 60; PMID: 10454532
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell 1998; 1:1021 - 31; http://dx.doi.org/10.1016/S1097-2765(00)80102-1; PMID: 9651585
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 1999; 13:1924 - 35; http://dx.doi.org/10.1101/gad.13.15.1924; PMID: 10444591
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol 1998; 8:843 - 6; http://dx.doi.org/10.1016/S0960-9822(98)70328-8; PMID: 9663395
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol 1998; 8:96 - 108; http://dx.doi.org/10.1016/S0960-9822(98)70040-5; PMID: 9427644
- Martínez-Balbás MA, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci U S A 1998; 95:132 - 7; http://dx.doi.org/10.1073/pnas.95.1.132; PMID: 9419341
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 2004; 14:183 - 93; http://dx.doi.org/10.1016/S1097-2765(04)00185-6; PMID: 15099518
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 2003; 130:4235 - 48; http://dx.doi.org/10.1242/dev.00625; PMID: 12900441
- Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol 2009; 53:275 - 89; http://dx.doi.org/10.1387/ijdb.082649rb; PMID: 19412887
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 2011; 25:755 - 66; http://dx.doi.org/10.1101/gad.2028911; PMID: 21460039
- Shuda K, Schindler K, Ma J, Schultz RM, Donovan PJ. Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev 2009; 76:1094 - 105; http://dx.doi.org/10.1002/mrd.21075; PMID: 19565641
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 1981; 84:145 - 58; http://dx.doi.org/10.1007/BF00293368; PMID: 7297248
- Robbins AR, Jablonski SA, Yen TJ, Yoda K, Robey R, Bates SE, Sackett DL. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle 2005; 4:717 - 26; http://dx.doi.org/10.4161/cc.4.5.1690; PMID: 15846093
- Choy JS, Acuña R, Au WC, Basrai MA. A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics 2011; 189:11 - 21; http://dx.doi.org/10.1534/genetics.111.130781; PMID: 21652526
- Vigneron S, Prieto S, Bernis C, Labbé JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom?. Mol Biol Cell 2004; 15:4584 - 96; http://dx.doi.org/10.1091/mbc.E04-01-0051; PMID: 15269280
- Jelluma N, Brenkman AB, van den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, Medema RH, Kops GJ. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell 2008; 132:233 - 46; http://dx.doi.org/10.1016/j.cell.2007.11.046; PMID: 18243099
- Murata-Hori M, Fumoto K, Fukuta Y, Iwasaki T, Kikuchi A, Tatsuka M, Hosoya H. Myosin II regulatory light chain as a novel substrate for AIM-1, an aurora/Ipl1p-related kinase from rat. J Biochem 2000; 128:903 - 7; http://dx.doi.org/10.1093/oxfordjournals.jbchem.a022840; PMID: 11098131
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 2007; 8:798 - 812; http://dx.doi.org/10.1038/nrm2257; PMID: 17848966
- Murata-Hori M, Wang YL. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr Biol 2002; 12:894 - 9; http://dx.doi.org/10.1016/S0960-9822(02)00848-5; PMID: 12062052
- Liu D, Lampson MA. Regulation of kinetochore-microtubule attachments by Aurora B kinase. Biochem Soc Trans 2009; 37:976 - 80; http://dx.doi.org/10.1042/BST0370976; PMID: 19754435
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 2004; 6:232 - 7; http://dx.doi.org/10.1038/ncb1102; PMID: 14767480
- Fadri-Moskwik M, Weiderhold KN, Deeraksa A, Chuang C, Pan J, Lin SH, Yu-Lee LY. Aurora B is regulated by acetylation/deacetylation during mitosis in prostate cancer cells. FASEB J 2012; 26:4057 - 67; http://dx.doi.org/10.1096/fj.12-206656; PMID: 22751009
- Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev 2006; 20:2566 - 79; http://dx.doi.org/10.1101/gad.1455006; PMID: 16980585
- Li X, Sakashita G, Matsuzaki H, Sugimoto K, Kimura K, Hanaoka F, Taniguchi H, Furukawa K, Urano T. Direct association with inner centromere protein (INCENP) activates the novel chromosomal passenger protein, Aurora-C. J Biol Chem 2004; 279:47201 - 11; http://dx.doi.org/10.1074/jbc.M403029200; PMID: 15316025
- Sasai K, Katayama H, Stenoien DL, Fujii S, Honda R, Kimura M, Okano Y, Tatsuka M, Suzuki F, Nigg EA, et al. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil Cytoskeleton 2004; 59:249 - 63; http://dx.doi.org/10.1002/cm.20039; PMID: 15499654
- Schindler K, Davydenko O, Fram B, Lampson MA, Schultz RM. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci U S A 2012; 109:E2215 - 22; http://dx.doi.org/10.1073/pnas.1120517109; PMID: 22778418
- Yang KT, Li SK, Chang CC, Tang CJ, Lin YN, Lee SC, Tang TK. Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol Biol Cell 2010; 21:2371 - 83; http://dx.doi.org/10.1091/mbc.E10-02-0170; PMID: 20484572
- Lane SI, Chang HY, Jennings PC, Jones KT. The Aurora kinase inhibitor ZM447439 accelerates first meiosis in mouse oocytes by overriding the spindle assembly checkpoint. Reproduction 2010; 140:521 - 30; http://dx.doi.org/10.1530/REP-10-0223; PMID: 20660090
- Sharif B, Na J, Lykke-Hartmann K, McLaughlin SH, Laue E, Glover DM, Zernicka-Goetz M. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci 2010; 123:4292 - 300; http://dx.doi.org/10.1242/jcs.067447; PMID: 21123620
- Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, Black BE. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol 2011; 21:1158 - 65; http://dx.doi.org/10.1016/j.cub.2011.06.015; PMID: 21723127
- Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J Biol Chem 2002; 277:27577 - 80; http://dx.doi.org/10.1074/jbc.C200307200; PMID: 12048181
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 2008; 16:1077 - 85; http://dx.doi.org/10.1016/j.str.2008.05.006; PMID: 18571423
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996; 272:408 - 11; http://dx.doi.org/10.1126/science.272.5260.408; PMID: 8602529
- Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem 2000; 275:9797 - 804; http://dx.doi.org/10.1074/jbc.275.13.9797; PMID: 10734134
- Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci U S A 2006; 103:7339 - 44; http://dx.doi.org/10.1073/pnas.0510946103; PMID: 16651529
- Yang F, Baumann C, Viveiros MM, De La Fuente R. Histone hyperacetylation during meiosis interferes with large-scale chromatin remodeling, axial chromatid condensation and sister chromatid separation in the mammalian oocyte. Int J Dev Biol 2012; 56:889 - 99; http://dx.doi.org/10.1387/ijdb.120246rd; PMID: 23417411
- Tang YA, Wen WL, Chang JW, Wei TT, Tan YH, Salunke S, Chen CT, Chen CS, Wang YC. A novel histone deacetylase inhibitor exhibits antitumor activity via apoptosis induction, F-actin disruption and gene acetylation in lung cancer. PLoS One 2010; 5:e12417; http://dx.doi.org/10.1371/journal.pone.0012417; PMID: 20856855
- Akiyoshi B, Nelson CR, Biggins S. The aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics 2013; 194:785 - 9; http://dx.doi.org/10.1534/genetics.113.150839; PMID: 23636741
- Schultz RM, Montgomery RR, Belanoff JR. Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol 1983; 97:264 - 73; http://dx.doi.org/10.1016/0012-1606(83)90085-4; PMID: 6189752
- Tsafriri A, Chun SY, Zhang R, Hsueh AJ, Conti M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol 1996; 178:393 - 402; http://dx.doi.org/10.1006/dbio.1996.0226; PMID: 8812137
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86:679 - 88; http://dx.doi.org/10.1530/jrf.0.0860679; PMID: 2760894
- Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol 2005; 7:93 - 8; http://dx.doi.org/10.1038/ncb1208; PMID: 15592459
- Duncan FE, Chiang T, Schultz RM, Lampson MA. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod 2009; 81:768 - 76; http://dx.doi.org/10.1095/biolreprod.109.077909; PMID: 19553597
- Stein P, Schindler K. Mouse oocyte microinjection, maturation and ploidy assessment. J Vis Exp 2011; http://dx.doi.org/10.3791/2851; PMID: 21808228
- Diaz-Rodríguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci U S A 2008; 105:16719 - 24; http://dx.doi.org/10.1073/pnas.0803504105; PMID: 18940925
