Abstract
Degradation of p53 is a cornerstone in the control of its functions as a tumor suppressor. This process is attributed to ubiquitin-dependent modification of p53. In addition to polyubiquitination, we found that p53 is targeted for degradation through ISGylation. Isg15, a ubiquitin-like protein, covalently modifies p53 at 2 sites in the N and C terminus, and ISGylated p53 can be degraded by the 20S proteasome. ISGylation primarily targets a misfolded, dominant-negative p53, and Isg15 deletion in normal cells results in suppression of p53 activity and functions. We propose that Isg15-dependent degradation of p53 represents an alternative mechanism of controlling p53 protein levels, and, thus, it is an attractive pathway for drug discovery.
Introduction
Controlling the stability of the p53 tumor suppressor is central to the regulation of its functions. This process has been intensively studied, and several E3 ligases, including the major ligase Mdm2, have been identified to target p53 through polyubiquitination for subsequent degradation by the 26S proteasome.Citation1-Citation4 Several recent reports, however, have provided evidence that proteasomal degradation of p53 can occur regardless of the status of p53 ubiquitination, and thus independent of Mdm2.Citation5-Citation7 These studies have led to an increased appreciation of the fact that p53 could also be degraded if newly synthesized p53 becomes intrinsically unstructured. There is no evidence to date that this degradation is an active process; however, different proteins have been proposed to bind the unstructured N and C terminus of p53, thereby potentially regulating 20S proteasome-mediated degradationCitation5,Citation7-Citation10.This mechanism is in contrast to the degradation of stabilized p53, which requires polyubiquitination to become susceptible to proteasomal degradation.Citation5
A recent study showed that the Isg15-modifying system is associated with the translational machinery and plays a role in broad targeting of newly synthesized proteins.Citation11 Isg15, the product of IFN-stimulated gene 15, was the first reported ubiquitin-like protein.Citation12 Isg15 is robustly induced by different stimuli, including type 1 IFNs, lipopolysaccharide, and viruses.Citation13-Citation16 Isg15 was also identified as a p53 target,Citation17 which is in line with a recent report that the chemotherapy-mediated induction of Isg15 requires functional p53.Citation18 Similar to ubiquitin conjugation, ISGylation of target proteins occurs in a 3-step cascade mechanism that involves Isg15-activating E1 enzyme (UBE1L), Isg15-conjugating E2 enzyme (UBCH8), and Isg15 E3 ligase with HERC5 being the main E3 ligase for Isg15.Citation16,Citation19-Citation22 More than 300 proteins are targeted for ISGylation;Citation20,Citation23-Citation26 however, only a handful of candidates have been validated as authentic substrates. In addition, the biological significance of this modification for many of the identified proteins remains largely unknown. Recent studies showed that ISGylation negatively regulates the ubiquitin–proteasome pathway by direct interference with polyubiqutination,Citation27 providing evidence of potential crosstalk between these 2 systems. Here we found that p53 is efficiently ISGylated by HERC5 and subsequently degraded by the 20S proteasome. Furthermore, Isg15 deletion increases the misfolded, dominant-negative p53, so we propose that ISGylation is like to work as a signal for degradation of misfolded p53, and this regulation is important for p53-mediated biological function.
Results
p53 is an Isg15 target protein
Because the Isg15-modifying system has been implicated in the broad targeting of newly synthesized proteins,Citation11 we investigated whether the tumor suppressor p53 was covalently modified by ISGylation. To do so, we transfected His-Isg15 together with Isg15-modifying E1 and E2 enzymes. Because it is unknown which E3 ligase targets p53 for ISGylation, we included several previously identified E3 ligases, including Herc5, as well as Efp and HhariI.Citation22,Citation28,Citation29 In addition, we included E3 ligase Mdm2 as well as MdmX as a negative control; this Mdm2 homolog does not have any E3 ligase activity. Isg15-modified proteins were pulled down with Ni beads and analyzed by western blotting with p53 antibody. We found that Herc5 efficiently modified p53 by ISGylation, with the appearance of 2 modified forms at approximately 68 and 83 kDa (). Based on the molecular weight of Isg15 (15 kDa), these 2 forms may correspond to p53 modified on 2 sites. Herc5 is a major Isg15-modifying E3 ligase,Citation20 and it has been shown to be associated with the translational machinery and to play a role in broad targeting of newly synthesized proteins, potentially including p53.Citation11 Because of this, for further analysis, we used Herc5 as the E3 ligase. First, we checked whether E3 ligase activity was required for p53 ISGylation by generating a Herc5 ligase-deficient mutant in which Cys994 was substituted for Ala.Citation20 We found that in contrast to wild-type Herc5, no p53 ISGylation was observed with the Herc5 C994A (). Usp18 acts as an Isg15-specific isopeptidase;Citation30 therefore, we next transfected cells with p53, the Isg15-modifying system (His-Isg15, E1, E2, and E3) and increasing concentrations of Usp18. We found that Usp18 was able to efficiently remove the Isg15 modification from p53 in a dose-dependent manner (). Thus, our data provide evidence that Herc5 efficiently modifies p53 by ISGylation, while Usp18 can remove this modification.
Figure 1. p53 is covalently modified by Isg15. (A) The E3 ligases Herc5 can modify p53 by ISGylation. HEK293T cells were transfected with p53, Ube1L (E1), UbcH8 (E2), and His-Isg15 or Myc-Isg15 (a negative control) together with different E3 ligases as shown in the panel. After Ni beads pulldown, Isg15-modified p53 was analyzed by western blotting with 1801 antibody. (B) The p53 ISGylation is dependent on the E3 ligase activity of Herc5. HEK293T cells were transfected with p53, Ube1L (E1), UbcH8 (E2), and His-Isg15 or Myc-Isg15 together with WT or ligase-dead (C994A) mutant of Herc5. Modified p53 was analyzed as in (A). (C) Usp18 decreases p53 ISGylation. Increasing concentration of Usp18 was transfected together with p53 and Isg15-modifying enzymes. Modified p53 was analyzed as in (A). (D) N-terminal and C-terminal region are required for efficient p53 ISGylation. The results of mass spectrometry analysis of ISGylated sites are shown in top panel, and identified lysines are indicated. Full-length or different truncated mutants of p53 as indicated in the panel were analyzed for ISGylation. (E and F) N-terminal lysine residues (K101, K120, K132, and K139) are required for p53 ISGylation. Different single (top panel) and multiple (bottom panel) point mutants of p53 were analyzed for ISGylation. The level of total p53 was used as a control.
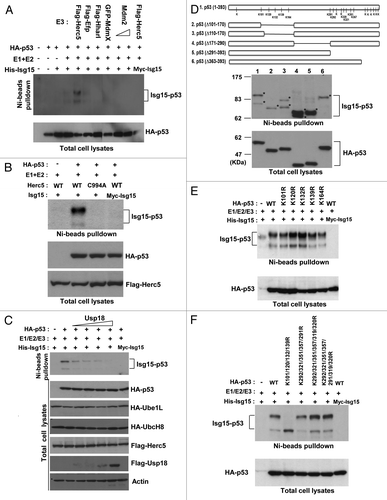
The Isg15-dependent modification of p53 results in the formation of 2 forms, with molecular weights of approximately 68 and 83 kDa (). Isg15 preferentially modifies lysine residues and, to a lesser extent, cysteines.Citation31 To verify whether cysteines are modified by Isg15, we incubated purified Isg15-modified p53 in the presence of a reducing agent, mercaptoethanol, and found that this was not sufficient to remove the modification (data not shown). Thus, we turned to the analysis of the lysine residues on p53. As there are 20 lysine residues that can be potentially modified on human p53, we first generated a series of truncated mutants to pinpoint the region of modification. As shown in , deletion of the N-terminal domain (amino acids 101–170) resulted in elimination of the top ISGylated band (; lane2), while deletion of the C-terminal domain (amino acids 291–393) resulted in loss of the second band (; lane 5). This supports the idea that one site is located in the N terminus, while the other is located in the C terminus of p53.
To identify the p53 ISGylation sites, the Isg15-modifying system (myc-Isg15, E1, E2, and E3) was transfected cells together with His-p53, and p53 was pulled down with Ni beads. We also co-transfected Arf to suppress Mdm2-dependent ubiquitination of p53, which may interfere with the analysis of Isg15-modified sites. After digestion with trypsin, we used a second immunoprecipitation with antibody recognizing a K-ε-GG motif with specificity for a di-glycine tag that is the remnant of Isg15 left on protein substrates after trypsin digestion (details in “Material and Methods”). Using this IP to enrich for Isg15-modified peptides and subsequent mass spectrometry, we found that on p53, 5 N-terminal (K101, K120, K132, K139, and K164) and 6 C-terminal lysine residues (K291, K292, K320, K321, K351, and K357) were modified when the Isg15 system was co-transfected with p53 (, top graph; Table S1). That multiple N- and C-terminal sites are modified by ISGylation based on mass spectrometry data while only 2 forms are present on the gel is not unexpected, because Isg15-dependent modifications in general are not very specific.Citation29 In addition, Isg15-dependent modification of a lysine residue covalently adds a substantial peptide structure (15kDa), which would prevent subsequent modification of other lysine residues within the same domain.Citation29 In this case, Isg15-dependent modification of any residue among the N-terminal sites would physically prevent modification of any other N-terminal residue within the same molecule. The same may apply to C-terminal sites. Thus, ISGylated p53 is most likely represented by a mixture of molecules modified at different sites in the N and C terminus.
To further verify ISGylated sites, single point mutations were generated in p53; these point mutants were then used for ISGylation analysis. We found that none of the single N-terminal mutants of the sites identified by mass spectrometry had a reduced level of p53 ISGlylation (). In contrast, mutation of 4 sites, K101R, K120R, K132R, and K139R, resulted in loss of the top ISGylated p53 band (). These data suggest that p53 could be equally modified on any of the 4 N-terminal sites, and modification of any single site, in turn, could potentially physically prevent subsequent modification of the neighboring sites.Citation29 Next, we analyzed the C-terminal lysine residues. Surprisingly, we found that mutation of all lysine residues identified by mass spectromentry was not sufficient to abrogate p53 ISGylation at the second site (). In addition to the 6 ISGylated C-terminal sites identified by mass spectrometry, there are 8 lysine residues in the C terminus of human p53. Based on our mass spectrometry data, it is most likely that physiologically only 6 C-terminal residues are modified by ISGylation in a wild-type sequence (). However, if these sites are mutated, additional C-terminal sites can be modified as well; a similar situation is described for p53 ubiquitination.Citation32,Citation33 Overall, our data indicate that Isg15-modified p53 comprises a mixture of different forms of p53 modified at 1 of the 4 (K101, K120, K132, and K139) N-terminal and 1 of the 6 (K291, K292, K320, K321, K351, and K357) C-terminal sites.
ISGylation targets p53 for proteasome-dependent degradation
We next examined the effect of ISGylation on p53. First, we checked whether ISGylation could change p53 localization. We found that regardless of expression of the Isg15-modifying system, p53 was nuclear (data not shown). Because ISGylation has been implicated in the broad targeting of newly synthesized proteins,Citation11 we investigated whether ISGylation targets p53 for proteasome-dependent degradation. To avoid the problem of different p53 levels between samples, we first transfected cells with saturating concentrations of p53. Under these conditions, when proteasome inhibitors cannot stabilize p53 further (lower panel, ), p53 ISGylation was markedly increased after treatment with the proteasome inhibitors MG132, epoxomicin, and lactacystin. Next we turned to the analysis of the effects of ISGylation on p53 at more physiological concentrations. We verified the specificity of our siRNA using an siRNA-resistant Isg15 (Fig. S1) and found that Isg15 knockdown increased the protein levels of p53 transfected at low concentrations as well as endogenous p53 (); in both cases, inclusion of the proteasome inhibitor diminished the difference. These data indicate that Isg15 may promote the proteasome-dependent degradation of p53.
Figure 2. p53 ISGylation promotes proteasome-dependent degradation. (A) Different proteasome inhibitors MG132 (25 μM), Epoxomicin (10 μM), and Lactocystin (10 μM) were used and p53 ISGylation was analyzed in HEK293T cells after transfecting p53 and Isg15-modifying system. (B) The Isg15 siRNA knockdown increases p53 protein levels. HEK293T cells were transfected with p53 and subsequently with Isg15 siRNA. p53 was visualized using HA antibody (HA-11).The knockdown efficiency of Isg15 was assessed by RT-PCR (bottom panel) (C). Similar to (B), but no p53 was transfected, and the level of endogenous p53 was analyzed with 1801 antibody in HCT116 cells. (D) ISGylated p53 can be degraded by proteasomes. Purified ISGylated p53 was incubated with 20S or 26S proteasomes and analyzed by western blotting using anti-p53 antibody 1801.
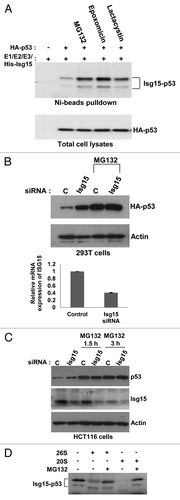
To determine if ISGylated p53 can be directly degraded by proteasomes, we incubated purified Isg15-modified p53 with different proteasome fractions in vitro. The in vitro degradation assay revealed that ISGylated p53 is degraded in the presence of 20S proteasomes and, to a lesser extent by the 26S proteasome (a complex of 19S and 20S subunits), a process that appeared to be sensitive to the proteasome inhibitor MG132 (). Together, these data suggest that Isg15 mediates the proteasome-dependent degradation of p53.
Ubiquitination and ISGylation cooperate in regulation of p53 levels
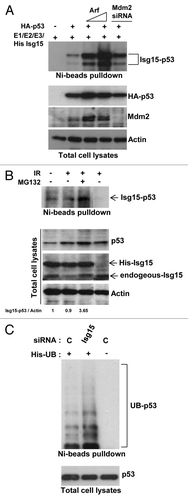
It is well established that polyubiquitination is a major pathway for p53 degradation. Polyubiquitination modifies lysine residues on p53, as does Isg15. Thus, these 2 types of modification could compete with each other for a lysine residue, yet the overall outcome would be similar: p53 degradation. Next, we examined the relationship between polyubiquitination and ISGylation of p53. Because p53 polyubiquitination is primarily regulated through Mdm2,Citation2,Citation3 we transfected cells with increasing concentrations of a negative regulator of Mdm2, Arf, or knocked down the level of Mdm2 with siRNA. p53 ISGylation was significantly enhanced when we blocked p53 polyubiquitination with Arf or Mdm2 siRNA (). We then treated HCT116 cells that express Flag-tagged endogenous p53 (obtained from T Waldman) with IR and MG132 and observed an increase in the ISGylation of endogenous p53 (). These data indicate that if polyubiqutination is blocked by IR or by downregulation of Mdm2 levels, the lysine residues may become available and thus could be modified by Isg15 to fine-tune the level of endogenous p53. This, in turn, suggests that both pathways could complement each other in controlling p53, and blocking them could have an additive effect. If the prediction is correct, and both ISGylation and polyubiquitination compete for the lysine residues, inhibition of ISGylation should increase p53 ubiquitination. Next, we transfected His-Ubiquitin, control, or Isg15 siRNA, and ubiquitinated proteins were precipitated with Ni beads, and endogenous p53 was analyzed. As shown in , p53 polyubiquitination was increased when the level of Isg15 was reduced with siRNA.
Figure 3. p53 ubiquitination and ISGylation together regulate p53 . (A) Arf overexpression or knockdown of Mdm2 increases ISGylation of p53. HEK293T cells were transfected with p53, Isg15-modifying enzymes, and increasing concentration of Arf or Mdm2 siRNA. After Ni beads pulldown, Isg15-modified p53 was analyzed by western blotting with 1801 antibody. (B) DNA damage together with a proteasome inhibitor enhances endogenous p53 ISGylation. HCT116 cells that contain Flag-tagged endogenous p53 (obtained from T Waldman) and expressing His-Isg15 were treated with IR (4 Gy) or MG132 (25 μM). Parental HCT116 cells were used as a control. After Ni-beads pulldown, Isg15-modified p53 was analyzed by western blotting with 1801 antibody. (C) Knockdown of Isg15 increases p53 ubiquitination. HEK293T cells were transfected with His-Ubiquitin, and control siRNA or Isg15 siRNA. After Ni beads pulldown, Ubiquitin-modified p53 was analyzed by western blotting with 1801 antibody.
ISGylation in normal cells targets misfolded p53
A recent study showed that the Isg15-modifying system is associated with the translational machinery and plays a role in broad targeting of newly synthesized proteins.Citation11 Thus, if p53 is not properly folded, it could be targeted by Isg15 for degradation. Deletion of Isg15 thus should result in accumulation of misfolded p53. To determine whether Isg15 regulates the level of p53 in different conformations, next we immunoprecipitated p53 with conformation-specific antibodies. The conformation of p53 can be assessed using Ab1620 antibody for wild-type p53Citation34 and Ab240 for p53 in the misfolded or denatured conformation.Citation35 We found that deletion of Isg15 in primary dermal fibroblasts (DF) resulted in significant (6-fold) accumulation of p53 in a misfolded conformation; the level of native p53 was either unchanged or slightly decreased (). This was accompanied by a decrease in the level of p53-responsive gene p21/Waf1 (), improved proliferation (), as well as attenuated replication-induced senescence of primary Isg15-deficient DF (). To understand the role of Isg15 in regulation of p53 in vivo, we next analyzed proliferation of neural stem/progenitor cells. It has been shown that their proliferation is declined rapidly in adult mice with p53 playing an important role,Citation36,Citation37 and we found that depletion of Isg15 resulted in increased proliferation of neural stem/progenitor cells ().
Figure 4. Knockout of Isg15 decreases p53 activity in dermal fibroblasts. (A) Isg15 knockout increases unfolding form of p53 in dermal fibroblasts (DF). Lysates from WT or Isg15 knockout DF were immunoprecipitated with p53 antibodies Ab1620 or Ab240. The immunoprecipitated p53 was analyzed by western blotting with CM-1 antibody. (B) Knockout of Isg15 decreases the expression of p21/Waf1 gene in DF. RT-PCR was performed to analyze the p21/Waf1 expression in WT or Isg15 knockout DF. (C) Incorporation of Edu is increased in Isg15 knockout DF. Edu incorporation assay was performed with WT or Isg15 knockout DF. Positively stained cells were counted, and quantification is shown. (D) Isg15 knockout inhibits replication senescence-associated β-galactosidase (SA-β-Gal) activity. DF (WT or Isg15 knockout) were fixed and stained for SA-β-Gal activity. Positively stained cells were counted, and mean ± s.d. is shown.
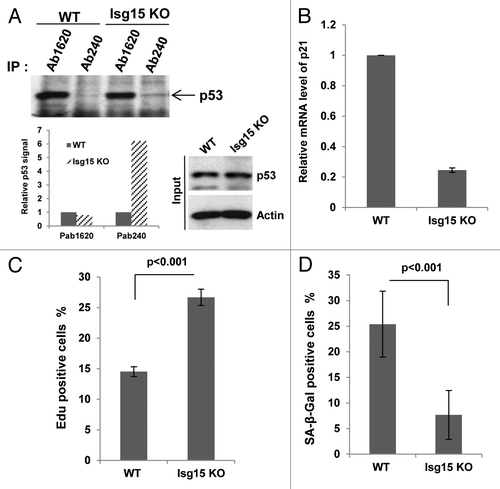
Figure 5. p53 activity is attenuated in Isg15 knockout mice. (A) Representative images of Edu labeling (right panels, red), counterstained with DAPI (left panels, blue) of WT (n = 3) and Isg15 knockout (n = 3) lateral ventricles. Note the specific Edu labeling in the sub-ventricular zone, where neural stem/progenitor cells are present. (B) Isg15 knockout increases unfolding form of p53 in thymocytes. Thymocytes collected from WT or Isg15 knockout mice were treated with IR (4 Gy) for 6 h before analyzing. p53 was immunoprecipitated using Ab1620 or Ab240 and analyzed by western blotting using CM-1 antibody. (C and D) Isg15 knockout decreases DNA damage-induced p53 activation and apoptosis. Thymocytes collected form WT (n = 5) or Isg15 knockout (n = 4) mice were treated with IR (4 Gy) and harvested at the indicated times. RT-PCR was performed to analyze the expression of p53 target genes (C). Annexin V-positive cells were analyzed by flow cytometry (D).
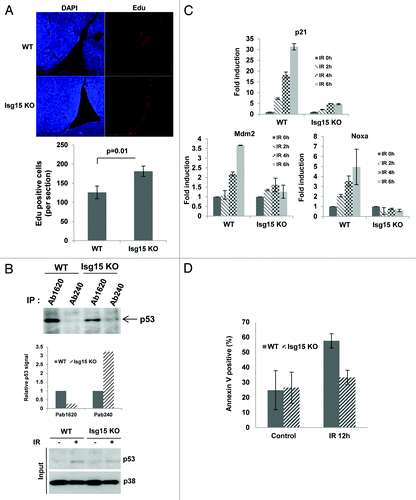
Deletion of Isg15 results in accumulation of misfolded p53, which normally has dominant-negative properties.Citation38 This, in turn, results in attenuation of p53 functions, including p53 response to DNA damage. To verify this, next we analyzed primary thymocytes and their response to 4 Gy of IR by assessing the activation of p53 target genes as well as a p53-dependent apoptosis.Citation39 Similar to DF, we found that deletion of Isg15 resulted in accumulation of misfolded p53 only (). Furthermore we observed a nearly complete abrogation of IR-induced activation of p21/Waf1, Mdm2, and Noxa in Isg15-deficient cells (). This attenuated p53-dependent transcription was accompanied by almost completely abrogation of apoptosis in Isg15-deficient thymocytes after IR (). Altogether our data argue that in normal cells, Isg15 primarily targets misfolded p53 for degradation and Isg15 deletion results in suppression of p53 activity and functions both in vitro and in vivo.
Discussion
It is widely believed that p53 is targeted for protein degradation solely through polyubiquitination. Several p53 E3 ligases have been identified, the major one of which is Mdm2.Citation1,Citation3,Citation4 The actual degradation process occurs in the 26S proteasome, which is composed of a core 20S catalytic unit and a 19S regulatory unit.Citation40 The role of 19S is in recognizing polyubiquitinated proteins, denaturing and redirecting them to 20S for degradation.Citation40,Citation41 It was believed that 19S must invariably fulfil its role in denaturing proteins before they can be degraded. However, a number of studies showed that certain proteins are naturally misfolded, and it was suggested that as much as 20% of all cellular proteins can be degraded or cleaved by the 20S proteasome directly.Citation42
In line with the fact that several unstructured proteins can be degraded directly by the 20S proteasome,Citation42 recent reports provide evidence for p53 proteasomal degradation regardless of its ubiquitination.Citation5,Citation7 Several proteins involved in the regulation of p53 degradation through the 20S proteasome have been proposed;Citation5,Citation8-Citation10 however, how unstructured p53 signals to 20S proteasome to initiate degradation was not clear. In our report, we argue that p53 ISGylation could be an overlooked type of covalent modification that provides a signal to target misfolded p53 for degradation by the 20S proteasome.
The biological functions of Isg15 conjugation remain largely unknown. It is believed that Isg15 may simply disrupt the function of the proteins to which it is conjugated.Citation26,Citation43 In our study, we argue that Isg15 may signal to other proteins or protein complexes in a manner similar to how ubiquitin signals to the proteasomes. We provide evidence that the majority of ISGylated p53 and components of the Isg15 system are present in fractions enriched for proteasomes () and, importantly, ISGylated p53 can be directly targeted by the 20S proteasome in vitro (). This function of Isg15 is not surprising, as previous studies have suggested that ISGylation may facilitate the proteasomal degradation of selected proteins.Citation44
In normal cells Isg15 is expressed at relatively low levels,Citation45-Citation47 and it was shown that Isg15-deficient mice have little phenotype.Citation48 More careful analysis of Isg15-deficient mice and cells derived from them, however, found that Isg15 is required for accumulation of misfolded p53, arguing that in normal cells ISGylation is an active process that primarily targets p53 in a misfolded conformation (). As this form of p53 is dominant-negative,Citation38 its accumulation after depleting Isg15 attenuates numerous aspects of a p53 response in normal cells. This includes p53’s role in the regulation of proliferation and replication-induced senescence in primary DF, proliferation of neural stem cells and progenitors, as well as IR-induced apoptosis of thymocytes (). Importantly, despite almost complete lack of apoptosis of Isg15-deficient thymocytes after IR, Isg15-knockout mice are not prone to develop tumors. These results are consistent with the fact that the pathological response to DNA damage does not contribute to p53-mediated tumor suppression,Citation49 while full transactivation of most p53 target genes is dispensable for p53 tumor suppressor function in a range of tissue types.Citation50
In summary, our study supports the hypothesis that ubiquitin-independent degradation of p53 is a significant process, and that Isg15, a protein that enhances this process, has an important role in maintaining the effective function of p53.
Materials and Methods
Animals, cell culture conditions, and treatments
All animal protocols used in this study were approved by the Institute of Molecular and Cell Biology Animal Safety and Use Committee. Isg15−/− mice were obtained from the Jackson Laboratory. Mouse embryo fibroblasts (MEFs) were purified from 12.5 d embryos.
HEK293T cells, HCT116 cells, and HCT116 Flag-tagged p53 cells were maintained in Dulbecco modified Eagle medium, and MCF7 cells in RPMI medium supplemented with 10% fetal bovine serum (Hyclone), 100 U/ml of penicillin, and 100 g/ml of streptomycin (Gibco). Cells were treated with Nutlin-3 (3 or 10 μM), or MG132 (25 μM), or Epoxomicin (10 μM), or Lactacystin (10 μM), or cycloheximide (20 μg/ml) and then harvested when appropriate.
To investigate p53 ISGylation of endogenous protein, HCT116 cells were transfected with expression construct encoding His-Isg15. The transfected cells were selected by using G418 (500 ug/ml) and were further screened for His-Isg15 expression by western blotting with anti-Isg15 antibody.
Plasmids and siRNA transfection
Plasmids expressing Mdm2 and hemagglutinin (HA)-tagged full-length p53 were gifts from Dr.Sheau-Yann Shieh (Institute of Biomedical Sciences). Plasmids expressing Myc-tagged Isg15, FLAG-tagged Herc5, HA-tagged Ube1L and UbcH8 were gifts from Dr Keh-Chuang Chin (Genome Institute of Singapore, Singapore). Plasmids expressing Efp, Hhari, Parkin, Nedd4, and SmurfF1 were obtained from Addgene. Full-length Isg15 was cloned into the BamH1 and Xba1 sites of pcDNA3.1/His vector (Invitrogen) or into the BamH1 and Xho1 sites of pXJ41-FLAG vector, a gift from Dr WJ Hong (Institute of Molecular and Cell Biology). To generate small interfering RNA (siRNA)-resistant Isg15-expressing constructs, conservative changes were made in the nucleotides targeted by Isg15 siRNA (5′-GGACAAGTGT GACGAACCT-3′; changes are underlined). All point and deletion mutants were generated by PCR-based site-directed mutagenesis. Plasmids were introduced into cells using calcium phosphate transfections.
Transfection of siRNA was performed using calcium phosphate precipitation in HEK293T cells and Oligofectamine (Invitrogen) in other cell lines. The following SMART pool siRNAs were purchased from Dharmacon: Isg15 (5′-GCAACGAAUU CCAGGUGUC-3′, 5′-GCAGAUCACC CAGAAGAUU-3′, 5′-GCACCGUGUU CAUGAAUCU-3′, 5′-GGACAAAUGC GACGAACCU-3′), Mdm2 (5′-GAUGAGAAGC AACAACAUA-3′, 5′-GAAUUUAGAC AACCUGAAA-3′, 5′-GAACAAGAGA CCCUGGUUA-3′, 5′-GCCAGUAUAU UAUGACUAA-3′).
Cell lysis, immunoprecipitation, and immunoblotting
Cells were lysed in TEGN buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 420 mM NaCl, 10% glycerol, and 0.5% Nonidet P-40) containing 1 mM dithiothreitol (DTT) and protease inhibitors cocktail (Roche). Antibodies used were the following: PAb1801 (sc98; Santa Cruz), p53 rabbit antibody (no. 9282; Cell Signaling), proteasome α3 (sc166205; Santa Cruz), proteasome β1 (sc58409; Santa Cruz), ISG15 (AP1150a; ABGENT), actin (A2066; Sigma), HA (16B12; Covance), FLAG M2 antibody (Sigma), and FLAG rabbit antibody (no.2368, Cell Signaling).
Analysis of the conformational form of p53 protein
p53 conformational form was analyzed by immunoprecipitation (IP). Cells were lysed in TEGN buffer containing protease inhibitors cocktail (Roche). For IP, the lysates were diluted with an equal volume of TEG and incubated with conformation-specific antibodies, PAb1620 (Calbiochem), or PAb240 (Calbiochem), and 15 μl of 50% protein G beads (Pierce) for 1.5 h at 4 °C. The immunoprecipitates were washed 3 times in IP buffer (TEGN: TEG = 1:1), and then boiled in a protein sample buffer and analyzed by SDS-PAGE (PAGE) followed by western blotting. The p53 in the immunoprecipitates was detected by p53 Sheep PAb (Ab-7) (Calbiochem).
Ni-beads pulldown assays
Transfected cells for different experiments were harvested and sonicated in buffer A (6 M guanidine-HCl, 0.1 MNa2HPO4/NaH2PO4, 10 mM imidazole, pH 8.0). Cell lysates were shaken with 30 μl of 50% Ni beads (Qiagen) at room temperature for 1 h. The precipitated beads were washed once in buffer A, once in buffer TI (20 mM imidazole, 0.2%Triton X-100, 25 mMTRIS-HCl, pH 6.8) mixed with buffer A (1:3), and 3 times in TI buffer. Proteins bound to beads were boiled in sample buffer and analyzed by SDS-PAGE followed by western blotting.
Two-step purification of Flag-ISG15-modified His-p53 and in vitro protein degradation assays
The HEK293T cells transfected with FLAG-Isg15, His-p53, HA-Ube1L, HA-UbcH8, FLAG-Herc5, and Arf at appropriate time points were re-suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 10 mM N-ethylmaleimide, proteases inhibitor cocktail (Roche), 1% Triton X-100, pH 8.0). After sonication and centrifugation, the lysates were incubated with Ni beads at 4 °C for 1 h. The precipitated beads were washed 3 times in wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 1% Triton X-100, pH 8.0). The His–p53 proteins bound to beads were eluted by elution buffer (50 mM NaH2PO4, 300 mMNaCl, 250 mM imidazole, 1% Triton X-100, pH 8.0), and dialyzed against PBS buffer with 10% glycerol. After dialysis, the His–p53 proteins were mixed with FLAG M2 beads, and incubated at 4 °C for 1 h. The precipitated beads were washed 3 times with 1:1 TEG/TEGN buffer. Finally, the FLAG-ISG15-modified His-p53 proteins were eluted by 3× FLAG peptides (Sigma), and then incubated with the 26S or 20S proteasomes (BostonBiochem) at 37 °C for 1.5 h. The reaction was stopped by the addition of a 0.5 volume of protein sample buffer, and the proteins were resolved by SDS-PAGE followed by western blotting.
EdU incorporation
DNA replication was measured using a Click-it EdU assay kit, which is based on incorporation of the thymidine analog 5-ethynyl-2’-deoxyuridine (EdU) into DNA during replication (Invitrogen). Then, 10 μM EdU was added to the cell culture medium 30 min before the cells were harvested and fixed in 4% paraformaldehyde. EdU was detected by flow cytometry after it reacted with Alexa647-azide (Invitrogen).
Apoptosis assay
To detect the phosphatidylserine externalization, thymocytes that collected form WT or Isg15 KO mice were double stained with FITC-conjugated Annexin-V and propodium iodide, according to manufacturer’s protocol (BD Biosciences). Annexin V-positive cells were collected by flow cytometry and analyzed using the CellQuest software (Becton Dickinson).
RT-PCR
Total RNA from the cells was extracted by TRIzol (Invitrogen), and the synthesis of cDNA was performed using an oligo(dT) primer by ReverAid™ Premium First Strand cDNA Synthesis Kit (Fermentas Life Science) according to the manufacturers’ protocols. The quantification of gene transcripts was analyzed by real-time PCR using SYBR green dye (KAPABIOSYSTEMS). All values were normalized to the expressing level of actin. The primers were as follows:
For human sample Isg15 forward, GAGAGGCAGC GAACTCATCT; Isg15 reverse, CTTCAGCTCT GACACCGACA; Actin forward, CCAGAGGAAG AGAGGCATCC; Actin reverse, GTGGTGGTGA AGCTGTAGCC. For mouse sample, p21 forward, AAGTGTGCCG TTGTCCTTC; p21 reverse, ACTTCAGGGT TTTCTCTTGC Mdm2 forward, AGGGGAAAGA TAAAGTGGAA; Mdm2 reverse, CCCCTGGCAG ATCACACATG; Noxa forward, AACGCGCCAG TGAACCCAAC; Noxa reverse, TGCCGTAAAT TCACTTTGTC; Actin forward, AGCCATGTAC GTAGCCATCC; Actin reverse, CTCTCAGCTG TGGTGGTGAA.
Mass spectrometry analysis of ISGylated sites
The eluates from Ni beads (eluted by imidazole and acid) were neutralized and pooled. The final volume was 2.7 mL. Reduction of cysteine disulfide bonds were performed by addition of 270 uL of 45 mM DTT to the cleared supernatant and incubating at 60 °C for 20 min. The solution was cooled to room temperature. Two hundred and seventy microliters (270 μL) of 110 mM chloroacetamide was added and incubated at room temperature in the dark for 15 min. The sample was diluted 4-fold. Forty μg of Trypsin was added, and the digestion was performed at 37 °C overnight. The resultant tryptic digest was cleaned up using a 3 mL C18 cartridge (3M) and vacuum dried.
The enrichment of the di-Gly-Lys-modified peptides was performed using ~20 μL PTMScan® K-GG Antibody Bead Conjugate (Cell Signaling Technology) following the supplier’s recommended protocol. The enriched modified peptides were stored on a self-packed stage tip made from 4 small C18 Empore disks (3M) packed into a 200 μL pipette tipsCitation51 before they were eluted and vacuum dried for LC/MS analysis.
Vacuum-dried sample was reconstituted in 0.1% formic acid and analyzed using nanoHPLC coupled to a LTQ Orbitrap classic (Thermo Fisher Scientific). Peptides were trapped onto a C18 pre-column and separated on an analytical column using a 4 h gradient ranging from 0 to 40% acetonitrile/0.1% formic acid, followed by a 5 min gradient ranging from 40–80% acetonitrile/0.1% formic acid. Survey full scan MS spectra (m/z 310–1400) were acquired with a resolution of r = 60,000 at m/z 400, an AGC target of 1e6, and a maximum injection time of 1000 ms. The 10 most intense peptide ions in each survey scan with an ion intensity of >2000 counts and a charge state ≥2 were isolated sequentially to a target value of 5000 and fragmented in the linear ion trap by collisionally induced dissociation using a normalized collision energy of 35%. A dynamic exclusion was applied using a maximum exclusion list of 500 with one repeat count, repeat duration of 45 s, and exclusion duration of 30 s.
Raw data files were analyzed using MaxQuant software (version 1.1.1.25) as described.Citation52 Database searches were performed with tryptic specificity allowing a maximum of 2 missed cleavages as well as an initial mass tolerance of 7 ppm for precursor ions and 0.5 Da for fragment ions. Cysteine carbamidomethylation was searched as a fixed modification, and N-acetylation, oxidized methionine, and GlyGly (K) were searched as variable modifications. Maximum false discovery rates were set to 0.01 for both protein and peptide. The GlyGly (K) modified peptides found on p53 were manually validated and shown in Table S1.
Statistical analysis
Values are mean +/− SEM. Comparison of mean values between group was evaluated by 2-tailed Student t test or ANOVA where indicated. A P value less than 0.05 was considered significant.
Additional material
Download Zip (65.4 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The research for D.V.B. and D.L. was supported by the Agency for Science, Technology and Research (Singapore). We are extremely grateful to T Waldman (The Georgetown University, USA) for providing HCT116 cells with Flag-tagged endogenous p53. We also thank Dr SY Shieh for providing us with the plasmids expressing MDM2 and HA-tagged full-length p53 and Dr KC Chin for Myc-tagged Isg15, FLAG-tagged Herc5, HA-tagged Ube1l and Ubch8.
References
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429:86 - 92; http://dx.doi.org/10.1038/nature02514; PMID: 15103385
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387:296 - 9; http://dx.doi.org/10.1038/387296a0; PMID: 9153395
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997; 387:299 - 303; http://dx.doi.org/10.1038/387299a0; PMID: 9153396
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003; 112:779 - 91; http://dx.doi.org/10.1016/S0092-8674(03)00193-4; PMID: 12654245
- Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A 2002; 99:3099 - 104; http://dx.doi.org/10.1073/pnas.052706799; PMID: 11867746
- Moscovitz O, Tsvetkov P, Hazan N, Michaelevski I, Keisar H, Ben-Nissan G, Shaul Y, Sharon M. A mutually inhibitory feedback loop between the 20S proteasome and its regulator, NQO1. Mol Cell 2012; 47:76 - 86; PMID: 22793692
- Tsvetkov P, Reuven N, Shaul Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ 2010; 17:103 - 8; http://dx.doi.org/10.1038/cdd.2009.67; PMID: 19557012
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 1998; 392:405 - 8; http://dx.doi.org/10.1038/32925; PMID: 9537326
- Maheswaran S, Englert C, Bennett P, Heinrich G, Haber DA. The WT1 gene product stabilizes p53 and inhibits p53-mediated apoptosis. Genes Dev 1995; 9:2143 - 56; http://dx.doi.org/10.1101/gad.9.17.2143; PMID: 7657166
- Nip J, Strom DK, Eischen CM, Cleveland JL, Zambetti GP, Hiebert SW. E2F-1 induces the stabilization of p53 but blocks p53-mediated transactivation. Oncogene 2001; 20:910 - 20; http://dx.doi.org/10.1038/sj.onc.1204171; PMID: 11314026
- Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell 2010; 38:722 - 32; http://dx.doi.org/10.1016/j.molcel.2010.05.002; PMID: 20542004
- Korant BD, Blomstrom DC, Jonak GJ, Knight E Jr.. Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J Biol Chem 1984; 259:14835 - 9; PMID: 6501319
- Kim MJ, Latham AG, Krug RM. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc Natl Acad Sci U S A 2002; 99:10096 - 101; http://dx.doi.org/10.1073/pnas.152327499; PMID: 12114540
- Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem 1992; 267:7806 - 13; PMID: 1373138
- Malakhova O, Malakhov M, Hetherington C, Zhang DE. Lipopolysaccharide activates the expression of ISG15-specific protease UBP43 via interferon regulatory factor 3. J Biol Chem 2002; 277:14703 - 11; http://dx.doi.org/10.1074/jbc.M111527200; PMID: 11854279
- Yang P, Yu Z, Gandahi JA, Bian X, Wu L, Liu Y, Zhang L, Zhang Q, Chen Q. The identification of c-Kit-positive cells in the intestine of chicken. Poult Sci 2012; 91:2264 - 9; http://dx.doi.org/10.3382/ps.2011-02076; PMID: 22912461
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature 1997; 389:300 - 5; http://dx.doi.org/10.1038/38525; PMID: 9305847
- Liu M, Hummer BT, Li X, Hassel BA. Camptothecin induces the ubiquitin-like protein, ISG15, and enhances ISG15 conjugation in response to interferon. J Interferon Cytokine Res 2004; 24:647 - 54; http://dx.doi.org/10.1089/jir.2004.24.647; PMID: 15684817
- Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol 2004; 24:9592 - 600; http://dx.doi.org/10.1128/MCB.24.21.9592-9600.2004; PMID: 15485925
- Wong JJ, Pung YF, Sze NS, Chin KC. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci U S A 2006; 103:10735 - 40; http://dx.doi.org/10.1073/pnas.0600397103; PMID: 16815975
- Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci U S A 2004; 101:7578 - 82; http://dx.doi.org/10.1073/pnas.0402528101; PMID: 15131269
- Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem 2006; 281:3989 - 94; http://dx.doi.org/10.1074/jbc.M510787200; PMID: 16352599
- Giannakopoulos NV, Luo JK, Papov V, Zou W, Lenschow DJ, Jacobs BS, Borden EC, Li J, Virgin HW, Zhang DE. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun 2005; 336:496 - 506; http://dx.doi.org/10.1016/j.bbrc.2005.08.132; PMID: 16139798
- Malakhov MP, Kim KI, Malakhova OA, Jacobs BS, Borden EC, Zhang DE. High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J Biol Chem 2003; 278:16608 - 13; http://dx.doi.org/10.1074/jbc.M208435200; PMID: 12582176
- Takeuchi T, Kobayashi T, Tamura S, Yokosawa H. Negative regulation of protein phosphatase 2Cbeta by ISG15 conjugation. FEBS Lett 2006; 580:4521 - 6; http://dx.doi.org/10.1016/j.febslet.2006.07.032; PMID: 16872604
- Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci U S A 2005; 102:10200 - 5; http://dx.doi.org/10.1073/pnas.0504754102; PMID: 16009940
- Desai SD, Haas AL, Wood LM, Tsai YC, Pestka S, Rubin EH, Saleem A, Nur-E-Kamal A, Liu LF. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res 2006; 66:921 - 8; http://dx.doi.org/10.1158/0008-5472.CAN-05-1123; PMID: 16424026
- Dastur A, Beaudenon S, Kelley M, Krug RM, Huibregtse JM. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem 2006; 281:4334 - 8; http://dx.doi.org/10.1074/jbc.M512830200; PMID: 16407192
- Okumura F, Zou W, Zhang DE. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev 2007; 21:255 - 60; http://dx.doi.org/10.1101/gad.1521607; PMID: 17289916
- Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem 2002; 277:9976 - 81; http://dx.doi.org/10.1074/jbc.M109078200; PMID: 11788588
- Bade VN, Nickels J, Keusekotten K, Praefcke GJ. Covalent protein modification with ISG15 via a conserved cysteine in the hinge region. PLoS One 2012; 7:e38294; http://dx.doi.org/10.1371/journal.pone.0038294; PMID: 22693631
- David Y, Ternette N, Edelmann MJ, Ziv T, Gayer B, Sertchook R, Dadon Y, Kessler BM, Navon A. E3 ligases determine ubiquitination site and conjugate type by enforcing specificity on E2 enzymes. J Biol Chem 2011; 286:44104 - 15; http://dx.doi.org/10.1074/jbc.M111.234559; PMID: 21965653
- Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 2010; 17:86 - 92; http://dx.doi.org/10.1038/cdd.2009.77; PMID: 19543236
- Milner J, Cook A, Sheldon M. A new anti-p53 monoclonal antibody, previously reported to be directed against the large T antigen of simian virus 40. Oncogene 1987; 1:453 - 5; PMID: 3330789
- Gannon JV, Greaves R, Iggo R, Lane DP. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J 1990; 9:1595 - 602; PMID: 1691710
- Meletis K, Wirta V, Hede SM, Nistér M, Lundeberg J, Frisén J. p53 suppresses the self-renewal of adult neural stem cells. Development 2006; 133:363 - 9; http://dx.doi.org/10.1242/dev.02208; PMID: 16368933
- Zhu YH, Zhang CW, Lu L, Demidov ON, Sun L, Yang L, Bulavin DV, Xiao ZC. Wip1 regulates the generation of new neural cells in the adult olfactory bulb through p53-dependent cell cycle control. Stem Cells 2009; 27:1433 - 42; http://dx.doi.org/10.1002/stem.65; PMID: 19489034
- Trinidad AG, Muller PA, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, Vousden KH. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell 2013; 50:805 - 17; http://dx.doi.org/10.1016/j.molcel.2013.05.002; PMID: 23747015
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie AH. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 1993; 362:849 - 52; http://dx.doi.org/10.1038/362849a0; PMID: 8479523
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67:425 - 79; http://dx.doi.org/10.1146/annurev.biochem.67.1.425; PMID: 9759494
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82:373 - 428; PMID: 11917093
- Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol 2009; 386:814 - 27; http://dx.doi.org/10.1016/j.jmb.2008.12.081; PMID: 19162040
- Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, Ka SH, Oh KH, Kim KI, Zhang DE, Bang OS, et al. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep 2009; 10:374 - 80; http://dx.doi.org/10.1038/embor.2009.23; PMID: 19270716
- Pitha-Rowe I, Hassel BA, Dmitrovsky E. Involvement of UBE1L in ISG15 conjugation during retinoid-induced differentiation of acute promyelocytic leukemia. J Biol Chem 2004; 279:18178 - 87; http://dx.doi.org/10.1074/jbc.M309259200; PMID: 14976209
- Guerra S, Cáceres A, Knobeloch KP, Horak I, Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog 2008; 4:e1000096; http://dx.doi.org/10.1371/journal.ppat.1000096; PMID: 18604270
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A 2006; 103:6001 - 6; http://dx.doi.org/10.1073/pnas.0601523103; PMID: 16585524
- Nicholl MJ, Robinson LH, Preston CM. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J Gen Virol 2000; 81:2215 - 8; PMID: 10950979
- Osiak A, Utermöhlen O, Niendorf S, Horak I, Knobeloch KP. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol 2005; 25:6338 - 45; http://dx.doi.org/10.1128/MCB.25.15.6338-6345.2005; PMID: 16024773
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 2006; 443:214 - 7; http://dx.doi.org/10.1038/nature05077; PMID: 16957739
- Jiang D, Brady CA, Johnson TM, Lee EY, Park EJ, Scott MP, Attardi LD. Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proc Natl Acad Sci U S A 2011; 108:17123 - 8; http://dx.doi.org/10.1073/pnas.1111245108; PMID: 21969549
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007; 2:1896 - 906; http://dx.doi.org/10.1038/nprot.2007.261; PMID: 17703201
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011; 10:1794 - 805; http://dx.doi.org/10.1021/pr101065j; PMID: 21254760
