Nearly 2 m of DNA is so tightly packaged that it fits neatly within the nucleus of each cell of our body. As if this was not mind-blowing enough, the problem is intensified in the male germline, as the nucleus of a sperm cell is about 6 times smaller than that of a somatic cell. Sperm are highly specialized cells charged with the sole responsibility of delivering the paternal genome to offspring. Thus, future generations depend upon the faithful delivery device of the sperm. The haploid genome of the father needs to be intricately packaged so that it fits within the confines of the sperm nucleus, and also so the genome is protected from the harsh environment that the sperm encounters during its epic journey from the safety of the testes, through the female reproductive tract, to its ultimate destination, the receptive oocyte.
But how is this achieved? In mammals, spermatogonial stem cells of the testes multiply by repeated rounds of mitosis and differentiate into primary spermatocytes, which subsequently undergo meiosis and become haploid round spermatids. Post-meiotic round spermatids inherit a nucleosome-based haploid genome. During spermiogenesis, nucleosomes are globally destabilized and evicted, canonical histones are acetylated, removed, and replaced by histone variants and testis-specific small basic proteins called transition proteins, which are ultimately replaced by the sperm-specific basic proteins protamines. Thus, the paternal genome is transformed from its nucleosome-based structure into a tightly packed toroid structure of mature sperm.Citation1 The molecular mechanisms underlying this repackaging are poorly understood, as the process is currently understudied due to its complexity and the lack of an in vitro experimental system to study it. While chromatin remodelers are considered crucial, no chromatin remodeler had been characterized in the spermiogenesic process, and the mechanics of the unpacking/repacking equipment have been obscure. In a recent study, the chromatin remodeler Chd5 (Chromodomain helicase DNA binding protein 5) was heralded as a master regulator that orchestrates the faithful repackaging of the sperm genome.Citation2
Chd5 is a member of the CHD family of chromatin remodelers that has nucleosome-stimulated ATPase activity.Citation3 Chd5 expression is restricted to post-meiotic spermatids, peaking just as the most dramatic chromatin remodeling starts to take place. Deficiency of Chd5 in mice results in defective spermatid maturation and sperm chromatin compaction, leading to reduced sperm counts and motility, increased abnormal sperm morphology, and male infertility, consistent with the observation that low CHD5 expression correlates with spermatogenic defects in humans. The defective compaction of chromatin in sperm lacking Chd5 is due to a multifaceted disruption of the histone-to-protamine replacement process within developing spermatids. Chd5 deficiency compromises histone H4 hyperacetylation, a hallmark considered essential for initiating histone removal. Chd5 deficiency also disrupts expression of specific histone variants, key components that facilitate removal of core nucleosomal histones. Consistent with its nucleosome-stimulated ATPase activity, Chd5′s absence compromises nucleosome eviction that normally follows H4 hyperacetylation. In addition, nucleosome removal generates DNA supercoiling tension that is relieved through double-stranded breaks, which must be repaired to ensure genome integrity. Chd5 is crucial for this process, as its deficiency augments DNA damage in both developing spermatids and mature sperm, consistent with Chd5′s role in activating the DNA damage response in somatic cells. Chd5 deficiency also elevates the expression of both transition proteins and protamines, possibly to compensate for the faulty histone retention. Chd5 binds to the Protamine 1 gene and regulates its transcription, while in concert mediates levels of protamine 2, transition protein 1, and transition protein 2, mainly post-transcriptionally. These findings identify Chd5 as the first chromatin remodeler with an orchestrating role in the histone-to-protamine remodeling process during spermiogenesis. Chd5 controls the cascade of molecular events underlying histone removal and mediates the homeostasis of transition proteins and protamines. Deficiency of Chd5 leads to unwarranted retention of nucleosomal histones and augmented DNA damage, as well as aberrant accumulation of transition proteins and protamines in spermatids and sperm, abnormalities associated with male infertility in humans.Citation4,Citation5 The cascade of defects in H4 hyperacetylation, nucleosome eviction, and DNA damage repair during maturation of Chd5-deficient spermatids also provides functional evidence elucidating the sequential order of these events.
Chd5 contains multiple functional domains (ATPase, Helicase, PHD, chromodomains, SANT, and DNA binding motifs), which may permit its diverse functions during spermiogenesis. Chd5 might also mediate multiple processes by regulating expression of specific genes. RNA sequencing reveals that Chd5 deficiency affects expression of genes implicated in chromosome organization, DNA damage response, alternative splicing, ubiquitin conjugation, etc., but does not cause a major change in global gene expression in spermatids. It is not yet clear whether Chd5 directly regulates these target genes. Questions also remain as to the identity of the specific loci that retain the abnormal nucleosomal histones and the consequence on the developmental potency of the sperm. Future studies that define the genome-wide distribution of Chd5 and how this impacts nucleosomal histones and specific histone modifications in developing spermatids and mature sperm should shed light on these questions. In addition, further investigation of CHD5′s role in human infertility might provide new avenues for better diagnosis and treatment for compromised male fertility. ()
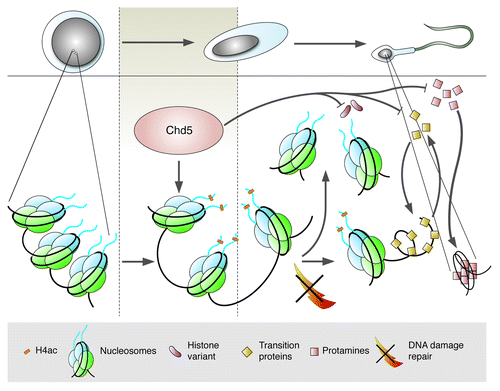
Figure 1. Chd5 regulates chromatin remodeling in sperm. Following meiosis, haploid spermatids develop into mature sperm (upper figure). Chromatin of round spermatids (left) contains canonical histones (blue, green) that are replaced with transition proteins (triangles) and ultimately protamines (squares). Chd5 expression (brown) in round/elongating spermatids precedes histone H4 hyperacetylation (H4ac, yellow rectangle), initiating eviction of nucleosomes, resolution of DNA supercoiling, and repair by the DNA damage response pathway (crossed lightning bolt). Loss of Chd5 compromises H4ac, eviction of nucleosomes, expression of histone variants/transition proteins/protamines, repair of DNA damage, culminating in jeopardized fertility. Schematic: Jim Duffy, Cold Spring Harbor Laboratory.
References
- Rousseaux S, et al. Reprod Biomed Online 2008; 16:492 - 503; http://dx.doi.org/10.1016/S1472-6483(10)60456-7; PMID: 18413057
- Li W, et al. Nat Commun 2014; 5:3812; PMID: 24818823
- Bergs JW, et al. PLoS One 2014; 9:e98203; http://dx.doi.org/10.1371/journal.pone.0098203; PMID: 24849318
- Ramos L, et al. Hum Reprod 2008; 23:259 - 70; http://dx.doi.org/10.1093/humrep/dem365; PMID: 18056059
- Oliva R. Hum Reprod Update 2006; 12:417 - 35; http://dx.doi.org/10.1093/humupd/dml009; PMID: 16581810
