ZMYND11 (also known as BS69), a candidate tumor suppressor that interacts with adenovirus E1A protein,Citation1 contains a tandem “reader” modules of histone modifications, including a plant homeodomain (PHD) zinc finger, a bromodomain and a PWWP domain. Recently, we found that this tandem PHD-bromo-PWWP domains of ZMYND11 recognize histone H3K36 trimethylation (H3K36me3) and importantly, this recognition is specific for the histone variant H3.3.Citation2 The PWWP domain is predominantly responsible for the readout of the trimethyl moiety, whereas a second composite pocket at the junction of the bromodomain, PWWP, and an embedded zinc finger motif recognizes the “Serine 31-Threonine 32 (S31-T32)” segment, thus accounting for the H3.3 specificity.
The PWWP domain belongs to the “Royal Family” of protein domains that also include Tudor, chromo, and MBT domains. It was initially identified as a DNA-binding domain,Citation3 whereas recently it has been shown that the PWWP domain, like other “Royal Family” members, also possesses methyl-lysine recognition activity, especially for methylation on histone H3K36.Citation4 Notably, ZMYND11 distinguishes itself from these PWWP proteins in that it is a histone variant H3.3-specific reader of K36me3.
The PWWP domain adopts a 5-bladed β-barrel fold with an extended C-terminal α-helix (αP). Histone H3K36me3 is recognized by a conserved aromatic cage formed at the center of the PWWP β-barrel. The ZMYND11 bromodomain adopts a typical 4 α-helical bundle fold but lacks canonical key residues for acetyl-lysine recognition.Citation2 One unique feature of ZMYND11’s reader module is the newly identified “C2CH”-type zinc finger motif embedded between bromo and PWWP. It stabilizes the V-shaped compact fold of the bromo-PWWP cassette and forms a composite pocket for the H3.3S31 recognition. Moreover, the surface of ZMYND11 bromo-PWWP is electrostatically patchy, implying a co-recognition mechanism of histone H3K36me3 and DNA in nucleosomal context. In addition, although the structure of H3K36me3-bound ZMYND11 tandem PHD-bromo-PWWP domains has yet to be determined, our biochemical studies demonstrated that the PHD finger also facilitates ZMYND11 association with H3K36me3 both in vitro and in vivo.Citation2 It remains an interesting topic for future studies to explore the exact functions of the PHD finger and the bromodomain of ZMYND11 and their contributions to the combinatorial readout of H3.3K36me3 at the nucleosomal level in concert with the PWWP domain.
H3.3 is a conserved histone H3 variant that is structurally close to the canonical H3. It differs from canonical H3.2 and H3.1 at only 4 or 5 amino acids, respectively, with 3 or 4 located in the core histone fold regions and only one residue (S31) located in the N-terminal unstructured tail. The “A87...I89G90” residues located in the histone-fold domain account for particular properties of histone H3.3, such as nucleosome stability and chaperone recognitions.Citation5 Although H3.3S31 is phosphorylated during mitosis, the role of this modification or S31 itself in transcriptional regulation is currently unknown. To our knowledge, our study is the first to define a critical role of H3.3 S31 in substrate recognition. Substitution of H3.3 S31 with Alanine (A31) present in canonical H3.1 severely impacted the binding of ZMYND11 to the H3K36me3 peptide and histones in vitro and in cells, respectively,Citation2 suggesting that the H3.3-specific S31 plays an indispensable role in ZMYND11’s combinatorial recognition of H3.3K36me3.
Our ChIP-seq analysis revealed that in accordance with previous reports,Citation6 H3.3 is enriched in the transcribed regions in addition to regulatory elements such as promoters and enhancers, and gene body-enriched H3.3 correlates with the distribution of H3K36me3.Citation2 The combination of H3K36me3 and H3.3 establishes a unique epigenetic state that defines the genomic distribution of ZMYND11, offering a spatiotemporal control of gene expression for both normal and neoplastic growth.
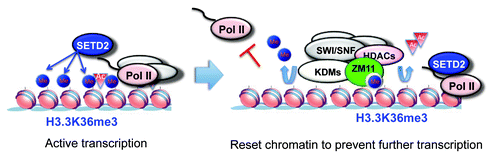
Although ZMYND11 is associated with genes with high H3K36me3 and expression levels, knockdown of ZMYND11 caused only a moderate change in gene expression.Citation2 We propose that ZMYND11, rather than working as an essential “on/off switch”, mainly functions to “fine-tune” gene expression. A similar “fine-tuning” mechanism has also been proposed for the actions of histone demethylases in transcriptional regulation. Interestingly, in a preliminary study, we found that ZMYND11 associates with several histone demethylases, histone deacetylases and the human SWI/SNF chromatin remodeler complex (Wen et al., unpublished data). It is tempting to speculate that ZMYND11 may regulate gene expression by modulating histone modifications and histone exchange in the transcribed regions, thus controlling the release of RNA Pol II from the pausing stage to the elongation stage (). Further work is required to answer whether ZMYND11 and its associated proteins function to reset chromatin after the passage of RNA Pol II, analogous to the better-understood processes in yeast modulated by the Isw1b and Eaf3/Rpd3 protein complexes.Citation7
Figure 1. Working model of ZMYND11 in resetting chromatin. During active transcription, SET2 associates with the elongating RNA Pol II and deposits trimethylation on H3K36 of histone H3.3. After the passage of RNA Pol II, ZMYND11 binds to the H3.3K36me3 and recruits histone demethylases (KDMs), histone deacetylases (HDACs), and the SWI/SNF chromatin-remodeling complex to reset the chromatin to a relatively repressive state to prevent further transcription.
References
- Hateboer G, et al. EMBO J 1995; 14:3159 - 69; PMID: 7621829
- Wen H, et al. Nature 2014; 508:263 - 8; http://dx.doi.org/10.1038/nature13045; PMID: 24590075
- Qiu C, et al. Nat Struct Biol 2002; 9:217 - 24; PMID: 11836534
- Wu H, et al. PLoS One 2011; 6:e18919; http://dx.doi.org/10.1371/journal.pone.0018919; PMID: 21720545
- Szenker E, et al. Cell Res 2011; 21:421 - 34; http://dx.doi.org/10.1038/cr.2011.14; PMID: 21263457
- Goldberg AD, et al. Cell 2010; 140:678 - 91; http://dx.doi.org/10.1016/j.cell.2010.01.003; PMID: 20211137
- Smolle M, et al. Epigenetics 2013; 8:10 - 5; http://dx.doi.org/10.4161/epi.23333; PMID: 23257840
