Abstract
The mechanisms that control E2F-1 activity are complex. We previously showed that Chk1 and Chk2 are required for E2F1 stabilization and p73 target gene induction following DNA damage. To gain further insight into the processes regulating E2F1 protein stability, we focused our investigation on the mechanisms responsible for regulating E2F1 turnover. Here we show that E2F1 is a substrate of the anaphase promoting complex or cyclosome (APC/C), a ubiquitin ligase that plays an important role in cell cycle progression. Ectopic expression of the APC/C activators Cdh1 and Cdc20 reduced the levels of co-expressed E2F-1 protein. Co-expression of DP1 with E2F1 blocked APC/C-induced E2F1 degradation, suggesting that the E2F1/DP1 heterodimer is protected from APC/C regulation. Following Cdc20 knockdown, E2F1 levels increased and remained stable in extracts over a time course, indicating that APC/CCdc20 is a primary regulator of E2F1 stability in vivo. Moreover, cell synchronization experiments showed that siRNA directed against Cdc20 induced an accumulation of E2F1 protein in prometaphase cells. These data suggest that APC/CCdc20 specifically targets E2F1 for degradation in early mitosis and reveal a novel mechanism for limiting free E2F1 levels in cells, failure of which may compromise cell survival and/or homeostasis.
Introduction
E2F1 belongs to the E2F family of transcription factors that regulate the expression of a wide variety of target genes important for cell cycle progression, DNA synthesis, apoptosis, DNA repair, mitosis and differentiation.Citation1–Citation3 Given that E2F1 has the ability to induce both cell cycle progression and apoptosis, aberrant expression of E2F can either promote or inhibit tumorigenesis depending on the cell type and conditions. This underscores the importance of ensuring that E2F1 levels and activity are tightly controlled throughout the cell cycle.
Multiple levels of regulation exist within the cell to control E2F1 activity. For example, E2F1 can induce its own transcription through the presence of E2F-responsive sites within its promoter.Citation4,Citation5 Post-transcriptionally, the DNA-binding activity of E2F1 is negatively regulated following phosphorylation by the cyclin A/cdk2 complex.Citation6 The regulation of E2F1 protein stability is another way by which E2F1 activity is controlled; however, the mechanisms behind this control remain poorly understood. E2F1 protein abundance can be regulated by the ubiquitin proteasome-dependent degradation pathway,Citation7–Citation9 and by stabilization through association with pRb, which protects E2F1 from ubiquitin-dependent degradation by binding to its C-terminal region.Citation9 The F-box protein p45Skp2 has been implicated in the ubiquitinmediated degradation of E2F1.Citation10 However, unlike other targets of Skp2, such as cyclin E and p27Kip1, E2F1 does not accumulate in Skp2-/- MEFs,Citation11 suggesting that Skp2 is dispensable for E2F1 degradation. E2F1 destruction can also occur in the nucleolar proteasome via interaction with ARF.Citation12 Furthermore, Mdm2 has been shown to affect E2F1 protein stability by inhibiting its ubiquitination.Citation13 E2F1 can also be ubiquitinated by multiple ROC-cullin ligases,Citation14 although the significance of this is not yet understood.
Upon DNA damage, E2F1 protein levels increase,Citation15,Citation16 and E2F1 induces apoptosis through the transactivation of various pro-apoptotic genes.Citation17 Two DNA damage-inducible phosphorylation sites have been reported within E2F1 that are required for its stabilization in response to DNA damage. ATM can modify serine 31 within the N terminusCitation18 and Chk2 phosphorylates serine 364 near the C terminus.Citation19 In addition, E2F1 acetylation at lysines 117, 120 and 125 has been shown to play a role in the activation and stabilization of E2F1 following DNA damage.Citation20,Citation21 It is unclear how these modifications cooperate to regulate E2F1 activity, and the disparate location of these sites within E2F1 suggests that the regulation of E2F1 stability is likely to be complex. We previously showed that the E2F1 transcription factor plays a key role in mediating the functional relationship between Chk1, Chk2 and p73 after genotoxic stress.Citation22 To gain further insight into how the protein stability and transcriptional activities of E2F1 are regulated following DNA damage, we initiated a series of experiments to better understand how the E2F1 protein itself is regulated throughout the cell cycle.
Here we show a role for the anaphase-promoting complex or cyclosome (APC/C), a large multiprotein E3 ubiquitin ligase, in regulating E2F1 protein stability. We find that E2F1 is targeted by APC/CCdc20 in vivo as a reduction in Cdc20 levels by RNA interference stabilizes the E2F1 protein and induces a specific accumulation of E2F1 in prometaphase-arrested cells. Thus, we have identified the APC/C as another key regulator of E2F1 activity within cells.
Results
Cdc20 and Cdh1 reduce ectopic E2F1 levels.
We first examined the expression of endogenous E2F1 using synchronized HeLa cell populations. HeLa cells were arrested at the G1/S boundary using a double thymidine block and then released into fresh media to allow cells to progress through the cell cycle. Whole cell extracts were prepared and E2F1 protein levels were analyzed by western blotting. We found that E2F1 levels were highest in cells arrested at the G1/S phase ( and B), and as cells progressed through S phase into G2/M, levels of E2F1 rapidly decreased. To further explore the kinetics of E2F1 regulation, we synchronized HeLa cells at prometaphase using a nocodazole block and followed E2F1 levels after release into fresh media. As shown in Figure 1C, E2F1 levels were undetectable at prometaphase and remained low as cells progressed through mitosis and G1. When cells reached the G1/S transition 10–12 h following release from the nocodazole block (), E2F1 levels peaked, consistent with the results from the double thymidine block. These data demonstrating that E2F1 is a cell cycle-regulated protein whose levels are highest at G1/S and are very low throughout mitosis are consistent with previous reports.Citation17
To gain further insight into the mechanisms regulating E2F1 protein stability, we wanted to investigate the involvement of ubiquitin ligases in E2F1 degradation. Given that E2F1 levels are regulated in a cell cycle-dependent manner, we decided to investigate a role for the SCF and APC/C complexes, two major E3 ligase complexes involved in cell cycle regulation, in degrading E2F1. Notably, the APC/C is most active when E2F1 levels are lowest during mitosis and G1. We tested whether E2F1 could be degraded by Skp2 and β-Trcp1, two substrate recognition F-box proteins of SCF, as well as the APC/C activators Cdc20 and Cdh1. HeLa cells were co-transfected with vectors encoding HA-E2F1 and HA-Cdc20, HA-Cdh1, FLAG-β-Trcp1 or FLAG-Skp2, and HA-E2F1 levels were examined by western blot. Interestingly, while ectopically expressed β-Trcp1 or Skp2 had a very mild, if any, effect on co-expressed E2F1 levels, E2F1 protein was dramatically decreased in the presence of Cdc20 or Cdh1. This result was also observed in T98G (), H1299 as well as U2OS cells (data not shown).
Next, we co-transfected HeLa cells with plasmids expressing E2F1 and Cdc20 or Cdh1 for 24 h, then treated cells with cycloheximide for 0, 1, 2, 4 or 6 h (). E2F1 was relatively stable when expressed alone (half-life greater than 6 h). However, in the presence of Cdc20 or Cdh1, the half-life of E2F1 was reduced to approximately 4 h, indicating that the Cdc20- and Cdh1- induced decrease in E2F1 abundance was due to an increase in E2F1 protein turnover. Note that the turnover of E2F1 from the time course using synchronized cells ( and B) shows that the physiological half-life of E2F1 is shorter than 4 hours. Therefore, the half-life of E2F1 measured here in asynchronous cells is probably an overestimate because of the mixing of mitotic cells with unstable E2F1 and interphase cells with stable E2F1. Taken together, these data suggest that the APC/C complex plays a role in ubiquitin-mediated turnover of E2F1.
DP1 partially protects E2F1 from APC/C-mediated degradation.
DP1 (a binding partner of the E2F family of transcription factors) and the retinoblastoma protein (pRb) are important regulators of E2F1 activity throughout the cell cycle. During G1/S, E2F1 is bound to DP1 as a heterodimer, and their association is critical for both the DNA binding and transcriptional activities of E2F1.Citation23,Citation24 In addition to protecting E2F1 from degradation in some settings, hypophosphorylated pRb binds to E2F1, and this complex acts as an active transcriptional repressor.Citation25 We therefore decided to investigate whether DP1 or pRb could interfere with E2F1 degradation by the APC/C. As shown in , E2F1 was degraded similarly by Cdc20 and Cdh1 in the presence or absence of pRb. However, E2F1 levels remained higher in cells co-transfected with DP1, suggesting that DP1 offers partial protection to E2F1 from degradation by Cdc20 and Cdh1. These results indicate that while pRb has no effect on APC/C-mediated E2F1 degradation, when E2F1 is bound as a heterodimer with DP1 the protein is unable to be efficiently targeted for degradation by Cdc20 and Cdh1.
E2F1 interacts with Cdc20 and Cdh1.
We next determined whether Cdc20 and Cdh1 could interact with E2F1. HA-tagged wild-type E2F1 and FLAG-tagged Cdc20, Cdh1 or a Cdc20 mutant with an N-terminal deletion (ΔN165) were expressed in HeLa cells, and immunoprecipitations for the FLAG-tagged proteins were performed to evaluate their interaction with E2F1. As shown in , E2F1 was readily detected in FLAG immunoprecipitates from cells expressing both E2F1 and Cdc20 or Cdh1, but not from cells expressing E2F1, Cdc20 or Cdh1 alone. Moreover, Cdc20 ΔN165 was able to co-immunoprecipitate E2F1. Cdc20 ΔN165 lacks the first 165 amino acids that contain a C-box, required to interact with APC/C core subunits, a KEN box and part of a CRY-box (both degradation motifs) and the Mad2-binding motif.Citation26 Although Cdc20 ΔN165 could bind to E2F1, this mutant could not degrade E2F1 in contrast to wild-type Cdc20 (). Amador et al. similarly showed that Cdc20 ΔN165 could bind to p21, a substrate of APC/CCdc20, yet was unable to target p21 for degradation.Citation26 Presumably, this is because Cdc20 ΔN165 is unable to bind to the APC/C core subunits, thereby sequestering substrates instead of delivering them to the APC/C for degradation. Taken together, these data provide evidence for an association of E2F1 with Cdc20 and Cdh1. Moreover, the N terminus of Cdc20 is not required for binding to E2F1 but is required for E2F1 degradation.
The destruction box motifs in E2F1 are not required for its degradation by Cdc20 and Cdh1.
The APC/C recognizes three types of motifs in its substrates, the destruction box (D-box, RXXLXXXXN), the KEN-box (KEN) and the A-box. By sequence analysis, we identified 2 RxxL motifs in E2F1 located in the cyclin A binding domain that could function as potential D-boxes (). The first D-box (position 80–83 in human E2F1) is conserved in mice, while the second D-box (position 91–94 in human E2F1) is conserved among mouse, Xenopus and human E2F3 proteins. No other potential APC/C recognition motifs, such as KEN or A-boxes, were identified in E2F1. To test the importance of the D-box motifs for E2F1 destruction by the APC/C, we constructed E2F1 mutants in which the arginine and leucine residues of the first D-box (ΔDbox1), the second D-box (ΔDbox2) or both D-boxes (ΔDbox 1 + 2) were mutated to alanine residues and determined whether the protein levels of these E2F1 mutants were reduced in the presence of coexpressed Cdc20 or Cdh1. As shown in , the mutants were degraded similarly to wild-type E2F1 by Cdc20 and Cdh1, suggesting that the D-boxes have no function in regulating E2F1 destruction by the APC/C.
E2F1 is a substrate of APC/CCdc20 in vivo.
To confirm that the APC/C is required for the destruction of E2F1 in vivo, we used small interfering RNAs (siRNAs) to inhibit the activity of Cdc20 or Cdh1 in HeLa cells and analyzed the expression of endogenous E2F1 (). Interestingly, we observed that knockdown of Cdc20 resulted in a large increase in the amount of E2F1 detected, suggesting that E2F1 degradation is mediated by Cdc20. Supporting the observation that Cdc20 is involved in E2F1 degradation. When levels of other well-characterized APC/CCdh1 substrates, such as Cdc20Citation27 and cyclin B1,Citation28–Citation30 increased upon depletion of Cdh1, E2F1 levels were reduced in these cells. We believe this is because depletion of Cdh1 raises Cdc20 levels and this induces degradation of E2F1. The differences seen in E2F1 protein levels in the transfected cells were not due to a change in their cell cycle distribution, as knockdown of Cdc20 or Cdh1 did not alter the cell cycle profile of the cells as determined by FACS analysis (data not shown).
Degradation experiments in HeLa cell extracts provided further confirmation that APC/CCdc20 degrades E2F1 in vivo. Following a previously published approach,Citation31 whole cell extracts were prepared from control, Cdc20- or Cdh1-siRNA transfected cells, supplemented with ubiquitin and cycloheximide, and E2F1 levels were followed over 6 hours to determine E2F1 stability under these conditions. When Cdc20 was silenced, E2F1 levels were increased and were only modestly reduced in the extracts over the time course ( and B). By contrast, levels of E2F1 were reduced following Cdh1 knockdown (), and E2F1 was degraded at a faster rate compared to control transfected cells (). The reduced levels of E2F1 and increased turnover in cells depleted of Cdh1 may be attributed to increased Cdc20 levels in these cells (). Cyclin B, a known substrate of APC/C, was used as a positive control in this assay. Depletion of Cdh1 has been shown to stabilize cyclin B in SH-SY5Y neuroblastoma cells and 293T embryonic kidney cells.Citation32 Similarly, we observed that cyclin B was stabilized in our Cdh1-knockdown cells ( and B). These data suggest that the stability of E2F1 is under the control of the APC/CCdc20 pathway in vivo.
In an attempt to demonstrate that Cdc20 is able to stimulate ubiquitination of E2F1 in cells we went on to perform in vivo ubiquitination assays. These experiments rely on the ability of the co-transfected E3 to stimulate conjugation of tagged ubiquitin to the target protein. Quite surprisingly, just the co-expression of E2F1 with HA-ubiquitin caused maximal ubiquitination of E2F1. We performed these experiments under multiple sets of conditions, including varying amounts and tags of the transfected proteins, but were not able to see any further stimulation of ubiquitination of E2F1 upon co-expression of Cdc20. While this result does not directly support our hypothesis, it does highlight the fact that E2F1 is capable of being highly ubiquitinated in cells. A study of functional consequences of such extensive ubiquitination would undoubtedly provide valuable insight into the activity of E2F1.
Silencing of Cdc20, but not Cdh1, stabilizes E2F-1 in early mitosis.
In order to determine when E2F1 is actively degraded by the APC/C during the cell cycle, we depleted Cdc20 or Cdh1 using siRNA, synchronized the cells in prometaphase using nocodazole and followed E2F1 expression upon release from the block through to G1/S. As shown in , in prometaphase-arrested cells (0 h post-nocodazole release), while there was no detectable E2F1 expression in the control cells or Cdh1-silenced cells, when Cdc20 expression was reduced, an increase in E2F1 levels was detected (compare 0 h time point between Luc si and Cdc20 si). This effect was also observed with a second siRNA targeting Cdc20 (data not shown). However, E2F1 was still able to be degraded in these cells. Levels of E2F1 were reduced by 4 h post nocodazole release, suggesting that other E3 ligases, such as Cdh1, may also play a role in E2F1 degradation. Notably, E2F1 mRNA levels remained unchanged when Cdc20 was silenced in both asynchronous and prometaphase-arrested cells (), suggesting that the accumulation of E2F1 protein was not due to an increase in E2F1 transcription. Furthermore, Cdc20 knockdown in cells arrested at G1/S by double thymidine block did not lead to increased E2F1 levels (data not shown), indicating that Cdc20 specifically targets E2F1 for degradation in prometaphase. Taken together, these data suggest that APC/CCdc20 promotes the degradation of E2F1 in early mitosis.
Discussion
The APC/C is a master regulator of the cell cycle and degrades multiple substrates in a precise temporal order.
Herein, we have provided multiple lines of evidence that demonstrate that E2F1 protein levels are regulated by the APC/C. First, we showed that ectopically expressed E2F1 is targeted for degradation by co-expressed Cdc20 or Cdh1. Second, endogenous E2F1 levels were increased following Cdc20 knockdown and remained stable in extracts over a time course, showing that this increase was directly due to stabilization of the protein and not a change in the rate of protein synthesis. Finally, siRNA directed against Cdc20 induced an accumulation of E2F1 protein in prometaphase cells. Together, these observations provide new insights into the cell cycle regulation of E2F1.
The E2F1/DP1 complex is abundant only at the G1/S boundary, 1 and regulates the expression of genes necessary for progression through the S and G2/M phases.Citation33 In late S phase, the cyclin A/cdk2 complex phosphorylates both DP1 and E2F1. These phosphorylation events release DP1 from E2F1 and reduce the DNA binding ability of E2F1, leaving ‘free’ E2F1 to be degraded through the ubiquitin/proteasome pathway. We found that co-expression of DP1 with E2F1 blocked Cdc20- and Cdh1-induced E2F1 degradation, suggesting that the E2F1/DP1 complex is refractory to APC/C regulation and that the APC/C can only target the ‘free’ form of E2F1 for degradation. Here we propose that this ‘free’ E2F1 is specifically targeted by APC/CCdc20 in prometaphase. However, while we observed an accumulation of E2F1 following Cdc20 knockdown, E2F1 was still able to be degraded in these cells. APC/CCdc20 is active from prometaphase through to anaphase, while APC/CCdh1 targets substrates for degradation in late mitosis/early G1.Citation32 It is possible that once Cdc20 has been degraded by Cdh1, Cdh1 may keep ‘free’ E2F1 levels low until G1/S when Skp2 acts to maintain appropriate levels and activity of E2F1, thus ensuring the proper regulation of E2F1-dependent gene activity. Interestingly, both Skp2Citation34 and Cdc20,Citation33 are E2F1-regulated genes, therefore E2F1 may function as part of a regulatory circuit to control its own fate. These data suggest that E2F1 may be targeted by multiple E3 ligases, which may not be surprising given that stringent control of its expression is so important as a result of the diversity of E2F1 function in multiple cellular processes.
The accumulation of E2F1 following Cdc20 knockdown in both unperturbed cells () and in cells in which the spindle assembly checkpoint is activated and sustained by nocodazole () indicates that Cdc20 promotes E2F1 degradation irrespective of the spindle checkpoint. While the bulk of Cdc20 activity is inhibited by the spindle checkpoint to prevent degradation of substrates such as securin and cyclin B, a subpopulation of Cdc20 remains active and can degrade several proteins in prometaphase, including cyclin A,Citation35 HoxC10,Citation36 Nek2ACitation37 and p21.Citation26 There is remarkable variation between the primary destruction motifs of APC/C substrates, and it is not clear how Cdc20 distinguishes which substrates are to be degraded prior to inactivation of the spindle assembly checkpoint. In addition to its D-box, Nek2A has a KEN-box and novel destruction motif at its C terminus required for degradation by APC/CCdc20. HoxC10 contains two D-boxes that are indispensible for its degradation, while p21 destruction requires only a minimal D-box. On the other hand, mutating the N-terminal D-box of cyclin A does not prevent its destruction by APC/CCdc20. Similar to cyclin A, we showed that mutating the two N-terminal D-boxes of E2F1 had no effect on its degradation by the APC/C. Further analyses are required to identify which motif(s) within E2F1 are recognized by Cdc20 and Cdh1 to facilitate its destruction.
Collectively, our data suggest that the mechanism underlying the APC/C-induced decrease in E2F1 protein is post-transcriptional and is a result of increased protein turnover. We were unable to rescue E2F1 degradation by the APC/C with the proteasome inhibitors MG-132, LLnL or lactacystin (data not shown), but in the same experiment we could rescue APC/C-mediated degradation of p21 and Cdc20. In fact, we could not stabilize endogenous or ectopic E2F1 with proteasome inhibitors in multiple cells lines and instead found that E2F1 levels were downregulated following treatment of cells with proteasome inhibitors at both the protein and mRNA levels. This observation was also reported by Lim et al. in human osteosarcoma cells;Citation38 however, the mechanisms responsible for this downregulation are currently unknown.
In conclusion, APC/C-mediated destruction of E2F1 provides a novel mechanism for controlling E2F1 levels in the cell cycle. As mentioned earlier, our previous studies showed that Chk1 and Chk2 are required for E2F1 stabilization and p73 target gene induction following DNA damage. Therefore, it is of future interest to determine whether the APC/C regulates E2F1 stability upon DNA damage and what role, if any, the Chk kinases play in this process.
Experimental Procedures
Cell lines, synchronization and drug treatment.
HeLa cervical carcinoma cells and T98G glioblastoma cells obtained from ATCC were cultured in Dulbecco's modified Eagle's medium (DMEM) plus 10% FBS at 37°C with 5% CO2. Cells were synchronized at G1/S using a double thymidine block. Briefly, HeLa cells were treated with 2 mM thymidine (Sigma) for 19 h, washed with PBS and cultured in fresh media for 9 h. The cells were treated again with thymidine for 16 h and were released from the block by washing in PBS and replacing the media with fresh DMEM + 10% FBS. Synchronization at prometaphase with nocodazole was performed by incubating cells for 16 h with 330 nM nocodazole (Sigma). The protein synthesis inhibitor cycloheximide (Sigma) was dissolved in H2O.
Mammalian expression plasmids and transfection.
HA-E2F1 and HA-DP1 have been described previously.Citation22 The HA-tagged Cdh1, HA-tagged Cdc20, FLAG-tagged Cdh1, FLAG-tagged Cdc20 wild-type, FLAG-tagged Cdc20 ΔN165 and FLAG-tagged Skp2 plasmids were provided by M. Pagano (NYU School of Medicine, New York, NY). FLAG-tagged β-Trcp1 was provided by A. Abeliovich (Columbia University, New York, NY). pCMV-Rb plasmid was a kind gift of Dr. Bill Kaelin (Dana-Farber Cancer Institute, Boston, MA). E2F1 D-box mutants were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturers instructions. All constructs were confirmed by sequencing. Transient cell transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturers instructions.
Cell cycle analysis.
Cellular DNA content was assessed by propidium iodide (PI) staining as follows. Cells were harvested, washed twice with PBS, fixed in 50% ice-cold ethanol for 30 min on ice, washed twice again with PBS and resuspended in PBS containing 66 µg/mL PI (Sigma) and 100 µg/mL RNaseA (Sigma). Cells were analyzed by flow cytometry on a FACScan cytometer (BD Biosciences) using CellQuest software. The percentage of cells in each cell cycle phase was quantified using the ModFit program.
Western blot analysis and immunoprecipitations.
For immunoblotting, cells were washed once in PBS, resuspended in TEGN buffer (10 mM Tris at pH 7.5, 1 mM EDTA, 10% glycerol, 0.5% NP40, 400 mM NaCl, 1 mM DTT, 0.5 mM phenylmethylsulfonylfluoride and protease inhibitor mixture containing 1 M Benzamidine, 3 mg/mL Leupeptin, 100 mg/mL Bacitracin and 1 mg/mL α2-macroglobulin) and incubated on ice for 30 min. Lysates were then cleared by centrifugation at 13,000 rpm for 5 min. Total protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories), and equal amounts of protein were separated on 10% SDS polyacrylamide gels and transferred to nitrocellulose membranes (Protran, Schleicher and Schuell). Membranes were then blocked for 1 h in 5% skim milk in 0.5% tween-PBS (tPBS) and then probed with primary antibody overnight at 4°C in tPBS. After washing 3 × 7 min with tPBS, membranes were incubated for 1 h with secondary horseradish peroxidase-conjugated antibody (Sigma) in tPBS. Following 3 × 7 min washes in tPBS, proteins were visualized by enhanced chemiluminesence (Amersham Biosciences). For immunoprecipitations, cells were lysed in TEGN buffer and 300 µg of protein/sample was incubated with 0.5 µL anti-FLAG M2 antibody (Sigma) for 16 h at 4°C. 10 µl of a 50/50 protein A-Sepharose (GE Healthcare) slurry and 20 µl of a 50/50 protein G-Sepharose (GE Healthcare) slurry were added for an additional hour before washing extensively in TEGN buffer. Bound protein was eluted by boiling in SDS sample buffer and immunoprecipitates were analyzed by immunoblotting as described above.
Antibodies.
The following primary antibodies were used: anti-E2F1 (C20 or KH-95, 1:1,000, Santa Cruz Biotechnology), anti-HA (HA.11, 1:1,000, Covance), anti-Flag M2 (1:2,000, Sigma), anti-MYC (9E10, 1:500, Santa Cruz Biotechnology), anti-DP1 (TFD10, 1:1,000, BD Biosciences), anti-Rb (IF-8, 1:5, supernatant from hybridoma culture), anti-Cdc20 (H-175, 1:1,000, Santa Cruz Biotechnology), anti-Cdh1 (DH01, 1:1,000, NeoMarkers), anti-cyclin B (GNS1, 1:1,000, Santa Cruz Biotechnology) and anti-actin (1:5,000, Sigma).
RNA interference.
siRNA duplexes were synthesized by Qiagen. The siRNA oligonucleotide sequences for Cdh1 were: 5′-(UGA GAA GUC UCC CAG UCA G)dTdT-3′ (sense) and 5′-(CUG ACU GGG AGA CUU CUC A)dTdT-3′ (anti-sense). The siRNA sequences for Cdc20 were: 5′-(CGG CAG GAC UCC GGG CCG A)dTdT-3′ (sense) and 5′-(UCG GCC CGG AGU CCU GCC G)dTdT (anti-sense). The siRNA sequences for Luc were: 5′-(CUU ACG CUG AGU ACU UCG A)dTdT (sense) and 5′-(UCG AAG UAC UCA GCG UAA G)dTdT (anti-sense). For siRNA transfection, cells were transfected with 50 nM siRNA using DharmaFECT 1 siRNA transfection reagent (Dharmacon) according to the manufacturers instructions.
In vivo ubiquitination experiments.
H1299 cells were co-transfected either with plasmids encoding Myc-E2F1, HA-ubiquitin or FLAG-Cdc20 (wild type or mutants). At 24 h post-transfection cells were harvested and Myc-E2F1 was immunoprecipitated using anti-Myc antibody. Immunoprecipitates were subjected to SDS-PAGE, transferred to nitrocellulose and both immuonoprecipitates and whole-cell lysates were blotted with anti-HA antibody for HA-tagged ubiquitin.
Degradation assay.
siRNA-transfected cells were harvested, washed with PBS, lysed in SB buffer (25 mM HEPES pH 7.5, 1.5 mM MgCl2, 5 mM KCl, 1 mM dithiothreitol, 15 mM creatine phosphate, 2 mM ATP, 0.5 mM phenylmethylsulfonylfluoride and protease inhibitor mixture containing 1 M Benzamidine, 3 mg/mL Leupeptin, 100 mg/mL Bacitracin and 1 mg/mL α2 macroglobulin) and homogenized by freeze-thawing and passage through a needle. Extracts were then subjected to centrifugation (5 min at 5,000 rpm followed by 60 min at 13,000 rpm), and total protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories). Extracts (30 µg/sample) were supplemented with degradation cocktail [1.5 mg/mL ubiquitin (PK-ubiquitin, prepared as previously describedCitation39), 7.5 mM creatine phosphate, 1 mM ATP, 1 mM MgCl2 and 0.1 mg/mL cycloheximide] and incubated at room temperature. Aliquots were collected at 0, 1, 2, 4 and 6 h for immunoblotting as described above.
RNA preparation and quantitative real-time PCR.
Total RNA was extracted from HeLa cells using the Qiagen RNeasy kit according to the manufacturer's instructions. RNA was additionally treated with RNase-free DNase I (Qiagen) to remove any residual genomic DNA. First-strand cDNA synthesis was performed with the Superscript III Supermix for qRT-PCR kit (Invitrogen) according to the manufacturer's instructions. Each PCR was carried out in triplicate in a 20 µL volume using Power SYBR Green PCR Master Mix (Applied Biosystems) for 15 min at 95°C for initial denaturing, followed by 35 cycles of 95°C for 30 s and 60°C for 30 s in the ABI Prism 7300 Real-Time PCR System. The ribosomal gene L32 was used as the control gene. Values for each gene were normalized to the expression levels obtained for L32, and fold induction was calculated versus time 0 h. Each reaction was done in triplicate from at least two independent experiments. Primer sequences are available upon request.
Figures and Tables
Figure 1 E2F-1 is degraded in late S/G2 phase. (A and B) HeLa cells were synchronized in prometaphase with 330 nM nocodazole for 16 h, collected and replated in fresh medium. Cells were harvested at the indicated time points and processed for immunoblotting (A) or FACS analysis (B). (C and D) HeLa cells were synchronized at G1/S using a double thymidine block as described in Materials and Methods. Following release from the double thymidine block, cells were harvested at the indicated time points and processed for immunoblotting (C) or FACS analysis (D).
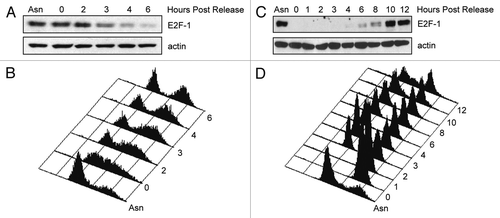
Figure 2 Cdc20 and Cdh1 reduce ectopic E2F-1 levels. (A) HeLa cells were transfected with HA-tagged E2F-1 alone and in the presence of HA-tagged Cdc20, HA-tagged Cdh1, FLAG-tagged β-Trcp1 or FLAG-tagged Skp2 at ratios of 1:5 and 1:15 (E2F1 and each ubiquitin ligase). All transfections were balanced with empty vector (EV, pcDNA3). Forty-eight hours after transfection, cells were harvested and extracts analyzed by immunoblotting with anti-HA, anti-FLAG or anti-actin antibodies. (B) T98G cells were transfected and analyzed as in (A). (C) HeLa cells were transfected with HA-tagged E2F-1 in the presence of HA-tagged Cdc20 (left part) or HA-tagged Cdh1 (right part) at a ratio of 1:15. Forty-eight hours after transfection, cells were treated with 100 µg/mL cycloheximide (CHX), collected at the indicated time points and extracts were analyzed by immunoblotting with anti-HA or anti-actin antibodies. Densitometric analysis was performed on E2F1 protein normalized to actin using Kodak 1D Image Analysis software. E2F1 alone was quantified using the lighter exposure and E2F1 co-expressed with Cdc20 or Cdh1 was quantified from the dark exposure.
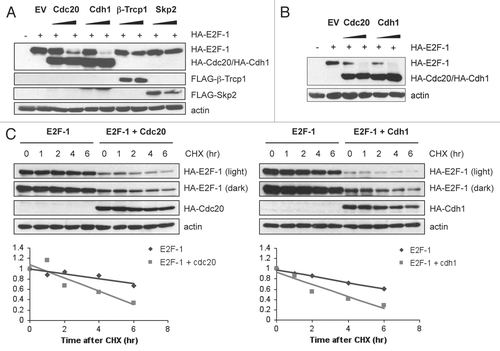
Figure 3 DP1, but not Rb, partially protects E2F-1 from APC/C-mediated degradation. (A) HeLa cells were transfected with HA-tagged E2F-1 (100 ng) in the presence of HA-tagged Cdc20 (1.5 µg), HA-tagged Cdh1 (1.5 µg), HA-tagged DP1 (100 ng) or pCMV-pRb (400 ng) as indicated. All transfections were balanced with empty vector (pcDNA3). 48 h after transfection, cells were harvested and extracts analyzed by immunoblotting.
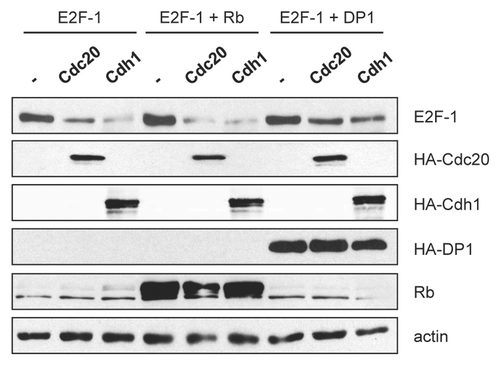
Figure 4 E2F1 interacts with Cdc20 and Cdh1. (A) HeLa cells were transfected with constructs encoding HA-tagged E2F1, FLAG-tagged Cdh1, Cdc20 or Cdc20 ΔN165 as indicated at a ratio of 1:1.5 (E2F1 and each ubiquitin ligase). All transfections were balanced with empty vector (pcDNA3). 24 h after transfection, cells were harvested and extracts were prepared. Equal amounts of protein were subjected to immunoprecipitation with anti-FLAG antibody as described in Materials and Methods, then analyzed by immunoblotting with anti-HA antibody. 10% of the lysate was used for the input samples. (B) HeLa cells were transfected with HA-tagged E2F-1 (100 ng) with increasing amounts of FLAG-tagged Cdc20 (wild-type), Cdc20 ΔN165 or Cdh1 (0, 0.5, 1 or 2 µg). All transfections were balanced with empty vector (pcDNA3). Forty-eight hours after transfection, cells were harvested and extracts analyzed by immunoblotting with anti-HA or anti-FLAG antibodies. (C) Alignment of the amino acid regions corresponding to the putative destruction box motifs (Dbox1 and Dbox2) in E2F1 with the D box motifs of cdc6, cyclin B1, securin, cyclin A, geminin, Skp2, Plk1 and p21. (D) HeLa cells were transfected with HA-tagged E2F-1 wild-type (wt) (100 ng), E2F-1 ΔDbox1 (100 ng), E2F-1 ΔDbox2 (100 ng) or E2F-1 ΔDbox1 + 2 (100 ng) alone or in the presence of HA-tagged Cdc20 (1.5 µg) or HA-tagged Cdh1 (1.5 µg). All transfections were balanced with empty vector (pcDNA3). Forty-eight hours after transfection, cells were harvested and extracts analyzed by immunoblotting with anti-HA or anti-actin antibodies.
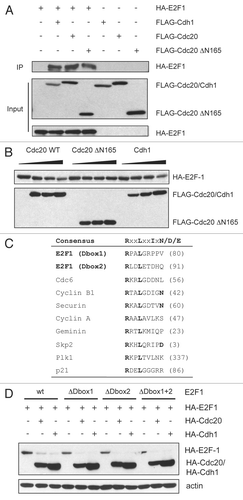
Figure 5 E2F1 is degraded in a Cdc20-dependent manner in vivo. (A) HeLa cells were transfected with control (Luc), Cdh1 or Cdc20 siRNA oligos. Forty-eight hours after transfection, cells were harvested and extracts analyzed by immunoblotting with antibodies to the indicated proteins. (B) Extracts prepared from cells treated as in (A) were supplemented with a degradation cocktail as described in Materials and Methods, incubated at room temperature and harvested at the indicated times. Levels of E2F1, cyclin B and actin were analyzed by immunoblotting. (C) E2F1 is hyper-ubiquitinated in cells. Extracts from H1299 cells transfected with Myc-E2F1 (100 ng), HA-Ubiquitin (500 ng), Flag-Cdc20 (50 and 100 ng) and Flag-Cdc20 ΔN (100 ng) as indicated, were immunoprecipitated (IP) with anti-Myc antibody as described in Materials and Methods. Ubiquitin conjugates were detected by immunoblotting IP samples with anti-HA antibody. Expression levels of transfected proteins were detected by western blotting shown in the right 3 parts (WCL).
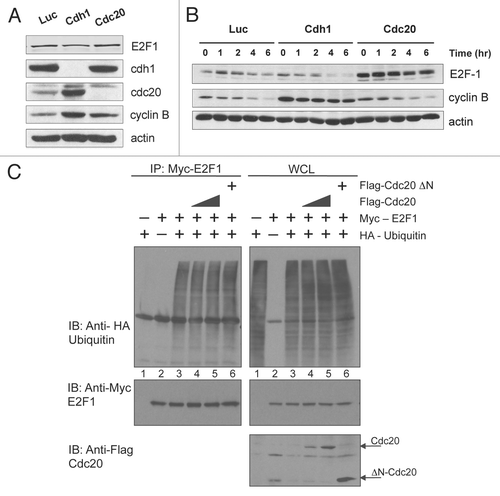
Figure 6 Cdc20 targets E2F1 for degradation in prometaphase. (A) HeLa cells were transfected with control (Luc), Cdc20 or Cdh1 siRNA oligos. 32 h later, fresh medium was added and cells were treated with 330 nM nocodazole for an additional 16 h to arrest cells in prometaphase. Cells were then replated in fresh media and harvested at the indicated times after release. Extracts were prepared and analyzed by immunoblotting with antibodies to the indicated proteins. (B) HeLa cells were transfected with control (Luc) or Cdc20 siRNA oligos. 32 h later, fresh medium was added and cells were left untreated (asynchronous) or treated with 330 nM nocodazole (Noc) for an additional 16 h to arrest cells in prometaphase. Total RNA was isolated and quantitative RT-PCR analysis was performed. Relative E2F1 mRNA levels were calculated by normalizing to L32 and data are representative of three independent experiments.
Acknowlegdements
We would like to thank the members of the Prives laboratory for their helpful input and discussions. This work was supported by grant #CA-87497 to C.P.
References
- Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ 1998; 9:585 - 593
- Stevens C, La Thangue NB. E2F and cell cycle control: a double-edged sword. Arch Biochem Biophys 2003; 412:157 - 169
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nature Rev 2002; 3:11 - 20
- Hsiao KM, McMahon SL, Farnham PJ. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev 1994; 8:1526 - 1537
- Neuman E, Flemington EK, Sellers WR, Kaelin WG Jr. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol 1994; 14:6607 - 6615
- Krek W, Ewen ME, Shirodkar S, Arany Z, Kaelin WG Jr, Livingston DM. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 1994; 78:161 - 172
- Campanero MR, Flemington EK. Regulation of E2F through ubiquitin-proteasome-dependent degradation: Stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA 1997; 94:2221 - 2226
- Hateboer G, Kerkhoven RM, Shvarts A, Bernards R, Beijersbergen RL. Degradation of E2F by the ubiquitin-proteasome pathway: Regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev 1996; 10:2960 - 2970
- Hofmann F, Martelli F, Livingston DM, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev 1996; 10:2949 - 2959
- Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSkp2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol 1999; 1:14 - 19
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J 2000; 19:2069 - 2081
- Martelli F, Hamilton T, Silver DP, Sharpless NE, Bardeesy N, Rokas M, et al. p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci USA 2001; 98:4455 - 4460
- Zhang Z, Wang H, Li M, Rayburn ER, Agrawal S, Zhang R. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene 2005; 24:7238 - 7247
- Ohta T, Xiong Y. Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases. Cancer Res 2001; 61:1347 - 1353
- Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol 1999; 19:3704 - 3713
- Hofferer M, Wirbelauer C, Humar B, Krek W. Increased levels of E2F-1-dependent DNA binding activity after UV- or gamma-irradiation. Nucleic Acids Res 1999; 27:491 - 495
- Tsantoulis PK, Gorgoulis VG. Involvement of E2F transcription factor family in cancer. Eur J Cancer 2005; 41:2403 - 2414
- Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev 2001; 15:1833 - 1844
- Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol 2003; 5:401 - 409
- Ianari A, Gallo R, Palma M, Alesse E, Gulino A. Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage. J Biol Chem 2004; 279:30830 - 30835
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol 2003; 5:552 - 558
- Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 2004; 18:3041 - 3054
- Bandara LR, Buck VM, Zamanian M, Johnston LH, La Thangue NB. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J 1993; 12:4317 - 4324
- Krek W, Livingston DM, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 1993; 262:1557 - 1560
- Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol 2007; 19:658 - 662
- Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell 2007; 27:462 - 473
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol 1998; 8:750 - 760
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 1995; 81:269 - 278
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 1995; 81:279 - 288
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell 1995; 6:185 - 197
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 2004; 432:588 - 595
- Almeida A, Bolanos JP, Moreno S. Cdh1/Hct1-APC is essential for the survival of postmitotic neurons. J Neurosci 2005; 25:8115 - 8121
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol 2001; 21:4684 - 4699
- Zhang L, Wang C. F-box protein Skp2: a novel transcriptional target of E2F. Oncogene 2006; 25:2615 - 2627
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol 2001; 153:137 - 148
- Gabellini D, Colaluca IN, Vodermaier HC, Biamonti G, Giacca M, Falaschi A, et al. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J 2003; 22:3715 - 3724
- Hayes MJ, Kimata Y, Wattam SL, Lindon C, Mao G, Yamano H, et al. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol 2006; 8:607 - 614
- Lim JH, Chang YC, Park YB, Park JW, Kwon TK. Transcriptional repression of E2F gene by proteasome inhibitors in human osteosarcoma cells. Biochem Biophys Res Commun 2004; 318:868 - 872
- Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 2007; 26:90 - 101