Abstract
Pura is a nucleic acid-binding protein with DNA-unwinding activity, which has recently been shown to have a role in the cellular response to DNA damage. We have investigated the function of Pura in Ultraviolet-C (UVC) radiation-induced DNA damage and nucleotide excision repair (NER). Mouse embryo fibroblasts from PURA-/- knockout mice, which lack Pura, showed enhanced sensitivity to UVC irradiation as assessed by assays for cell viability and clonogenicity compared to Pura positive control cultures. In reporter plasmid reactivation assays to measure the removal of DNA adducts induced in vitro by UVC, the Pura-negative cells were less efficient in DNA damage repair. Pura-negative cells were also more sensitive to UVC-induced DNA damage measured by Comet assay and showed a decreased ability to remove UVC-induced cyclobutane pyrimidine dimers. In wild-type mouse fibroblasts, expression of Pura is induced following S-phase checkpoint activation by UVC in a similar manner to the NER factor TFIIH. Moreover, co-immunoprecipitation experiments showed that Pura physically associates with TFIIH. Thus, Pura has a role in NER and the repair of UVC-induced DNA damage.
Introduction
Ultraviolet (UV) treatment of mammalian cells represents one of the most studied experimental systems for the examination of the cellular effects of DNA damage.Citation1 UV light induces the formation of covalent linkages between adjacent pyrimidine residues on the same DNA strand to give premutagenic lesions known as pyrimidine dimers. Pyrimidine dimers are pairs of thymine and cytosine bases that arise via photochemical reactions whereby UV light induces formation of covalent linkages between them. The two most common UV products are cyclobutane pyrimidine dimers (CPD) and the 6,4-pyrimidinepyrimidone photoproducts (6,4PP).Citation2 These premutagenic lesions alter the structure of DNA and consequently inhibit polymerases and arrest transcription and replication. DNA photoproducts are potentially mutagenic and lead to the activation of the S-phase checkpoint that can signal to activate p53-dependent DNA repairCitation3 and apoptosis.Citation1
Exposure of cells to UVC radiation is widely used as a model for studying the consequences of DNA damage and mechanisms of its repair because UVC light with wavelength ranging from 100 to 295 nm overlaps the absorption peak of DNA at 260 nm. Since UVC is not efficiently absorbed by proteins UVC damage is more specific for DNA.Citation4 UVC radiation is absorbed by the atmospheric ozone layer so that UVA and UVB are the main parts of solar UV that reach the surface of the earth. However, many of the DNA lesions induced by UVC are the same as the lesions induced by UVA and UVB.Citation4
In mammalian cells, the repair of damaged DNA containing UV photoproducts is mediated by the process of Nucleotide Excision Repair (NER) that involves about 30 proteins including the seven xeroderma pigmentosum (XP) genetic complementation group proteins, the DNA damage recognition complex XPChHR23B and the transcription/repair factor TFIIH. These bind sequentially to the DNA lesion and excise it.Citation5 There are two pathways for NER, a general pathway known as global genomic repair (GGR) and a transcription-coupled repair process (TCR), which is confined to DNA lesions that are in the transcribed strand of transcriptionally active genes and requires transcription by RNA polymerase II.Citation1 Defects in NER are associated with severe human diseases that involve abnormal sensitivity to sunlight including Xeroderma pigmentosum and Cockayne's syndrome, thus underscoring the importance of NER. Cells that are deficient in NER, e.g., XP cells, are extremely sensitive to UV light reflecting the power of unrepaired UV photoproducts to trigger apoptosis after UV irradiation (reviewed in ref. Citation1).
Our recent studies of the nucleic acid-binding protein Purα have revealed that it has a role in DNA repair including replication-associated DNA repair of double-strand breaks, homologous recombination-directed (HRR) DNA repair and the response of cells to DNA damage induced by cisplatin.Citation6–Citation9 Importantly, major lesions formed in DNA by cisplatin include intrastrand linkages between adjacent guanines. Thus, an upregulation of NER activity has been associated with cisplatin resistance in certain tumors.Citation10 Because NER is a major repair mechanism for DNA lesions induced by cisplatin and Purα has a protective role to cisplatin,Citation6 we have now investigated the role of Purα in NER and in the cellular response to UV irradiation.
Purα is a highly conserved cellular regulatory protein that was first isolated from mouse brainCitation11,Citation12 and human HeLa cells.Citation13,Citation14 Mouse PurαCitation15 differs from human Purα by only two amino acids,Citation14 and the Purα DNA-binding domain shows strong evolutionary conservation. Two orthologs of Purα have also been described: PurβCitation14 and Purγ.Citation16 Purα has a ubiquitous tissue distribution and is a multifunctional protein, which binds to both DNA and RNA and is thought to function in the initiation of DNA replication, control of transcription and mRNA translation.Citation9,Citation17,Citation18 Purα is also involved in the transport of mRNAs, including HIV-1 intron-containing mRNACitation19 and in the transport and targeting in the kinesin-associated granules of the dendrites of neuronal cellsCitation20,Citation21 and has been implicated in cell cycle regulation and oncogenic transformation since it binds to several cell cycle regulatory proteins, can cause cell cycle arrest and can inhibit the growth of tumor cells (reviewed in ref. Citation9).
Targeted inactivation in knockout mice of the PURA gene encoding Purα revealed that Purα has an essential role in postnatal brain development since PURA-/- mice develop neurological problems at two weeks of age and die by four weeks.Citation22 PURA-/- mice allow the preparation of primary cultures of mouse embryo fibroblast (MEFs) that are deficient for Purα and hence can be used as an experimental system to examine the cellular functions of Purα. Using this cell system, we have found that Purα can affect cellular DNA repair. Firstly, Purα has a role in the cellular response to replication-associated DNA repair of double-strand breaks since Purα negative cells are hypersensitive to the DNA replication inhibitor, hydroxyurea, suggesting a role for Puralpha as a caretaker protein that is involved in the repair of DSBs induced by stalled replication forks.Citation7 Secondly, Purα regulates homologous recombination-directed DNA repair (HRR) by modulating the expression of the HRR protein Rad51.Citation8 Finally, cells lacking Purα showed enhanced sensitivity to cisplatin-induced DNA damage and impaired non-homologous end-joining DNA repair.Citation6 In light of these findings, we have now investigated whether Purα may have a role in the cellular response to DNA damage induced by UV irradiation and NER.
Results
Purα-negative mouse embryo fibroblasts (Purα−) are more sensitive to UVC in cell viability and clonogenicity assays compared to Purα-positive mouse embryo fibroblasts (Purα+).
The effect of UVC on Purα+ and Purα− MEFs was examined (). Purα− cells were more sensitive to UVC and this was most pronounced at a dose of 10 J/m2 (). Similarly a time course for the effect of UVC showed that Purα-deficient MEFs exhibited higher sensitivity as early as 48 hours following irradiation (). The Purα status of the cells used in and B was confirmed by western blot (). Pretreatment of Purα+ cells with siRNA to Purα increased their sensitivity to UVC relative to controls treated with non-targeting siRNA () whereas ectopic expression of Purα in Purα− cells augmented their resistance to UVC (). Reduction of Purα expression by Purα siRNA in the experiment shown in was substantial as measured by western blot (). Expression of Purα from pCMV-Purα in was confirmed by western blot (). Thus Purα exerts a protective effect against UVC radiation. Similar results were obtained using a clonogenicity assay ().
Purα− cells are impaired in their ability to reactivate transfected UV-irradiated reporter plasmid compared to Purα+ cells.
This assay measured the ability of cells to repair DNA damage inflicted in vitro to a reporter plasmid by UVC irradiation. Purα+ and Purα− MEFs were transfected with luciferase reporter plasmid, which had been irradiated with various doses of UVC. Control cultures were transfected with plasmid that had not been irradiated. The activity of the reporter luciferase was determined as a measure of DNA repair capacity. Purα+ cells repaired and restored luciferase activity of irradiated plasmid more efficiently than Purα− cells (). Introduction of ectopic Purα into Purα− cells by transfection with pCMV-Purα improved DNA repair but pCMV vector did not (). Purα- cells transfected with pCMV-Purα repaired irradiated plasmid even more efficiently than Purα cells and this may be because they express higher levels of Purα protein than Purα cells as measured by western blot ( and compare lane 4 to lane 1). We conclude that Purα improves the repair of UVC-irradiated plasmid. Interestingly, because reactivation of UVC-damaged reporter plasmid involves repair of a transcriptionally active gene, this assay measures the capacity of the host cells to perform TCR, a pathway of NER that targets transcribed gene sequences.
Purα− cells show a decreased ability to remove UVC-induced cyclobutane pyrimidine dimers (CPDs) compared to Purα+ cells.
Pyrimidine dimers are produced by photochemical reactions induced by UV light and one of the most common UV products is CPDs.Citation2 In cells irradiated with UVC, the approximate ratio of CPD to (6-4)PP is 3:1.Citation23 To compare the occurrence of CPDs in Purα+ and Purα− MEFs in response to UVC irradiation, cells were analyzed by FACS with a monoclonal antibody that recognizes thymine dimers (). Thymine dimers were more prevalent in the Purα− cells than in the Purα+ cells. At 24 h after irradiation, 64.8% of the Purα− cells contained CPDs compared to 56.5% for the Purα+ cells (right hand panels, green peaks).
We analyzed the CPD formation kinetics using a slot blot method where DNA extracted from irradiated cells was spotted onto a nitrocellulose membrane and probed with antibody against CPDs (). The difference in the amount of CPDs between Purα+ and Purα− cell lines was significant at 24 h and increased greatly at 48 h after UV treatment suggesting that removal of UVC-induced pyrimidine dimers in the absence of Purα is less effective.
Purα− MEF s show increased DNA fragmentation in response to UVC irradiation in the comet assay compared to Purα+ cells.
Strand breaks observed after UV irradiation can be caused by interruption of replication at sites of DNA lesions, and also by processing of unrepaired lesions. Double-stranded breaks and single- stranded DNA breaks (nicks) can be detected using the Comet assay, where duplex cellular DNA is denatured and the fragmented DNA is resolved. Cells are embedded in agarose on a microscope slide, lysed and subjected to alkaline agarose gel electrophoresis. Intact DNA is revealed as a compact “comet” head and broken DNA as a “tail.” This provides a measure of cellular DNA fragmentation and its repair and allows the demonstration of the presence of single-strand breaks in the DNA of cells exposed to UV and the investigation of the kinetics of their repair.
Purα+ and Purα− MEFs were irradiated with UVC. Unirradiated cells served as control. The mean Olive Tail Moment (OTM), representing the product of the tail length and the fraction of the total DNA in the tail is used as a measure of DNA fragmentation (). As shown in , UVC caused an increase in the percent of cells with an OTM greater than the arbitrarily chosen value of 13. This indicates the presence of DNA fragmentation at the 48 h and 72 h time points. This increase was greater in the Purα− cells. Interestingly, the portion of the Purα− cells with OTM >13 have progressively increased from 26% at 48 h to 48% at 72 h (), while the percent of Purα+ cells with fragmented DNA did not change significantly over the time. We conclude that increased fragmentation of DNA observed in Purα− cells after UV irradiation results from inefficient removal of CPDs due to impaired NER. Thus Purα− cells are more prone to UVC-induced DNA fragmentation than Purα+ cells.
UVC irradiation has a differential effect on DNA damage signaling pathways in Purα+ and Purα− cells.
In order to examine cell cycle checkpoint signaling pathways, Purα+ and Purα− cells were synchronized in S-phase by double thymidine block, irradiated with UVC and were analyzed by western blot (). Six hours after UV irradiation, phosphorylation of H2AX, ATM, ATR and to a lesser extent p53 was significantly more enhanced in the Purα− cells compared to the Purα+ cells (compare lanes 2 and 4). After 24 hours, this difference in phosphorylation was still evident for ATM and ATR. In order to verify that the thymidine block and UV-mediated cell cycle arrest were occurring, the same cells were subject to FACS analysis (). At time zero, around 37–41% of cells were in S phase which dropped to approximately 22% in unirradiated cells after 24 hours but remained at around 40% in the UVC-irradiated cells. Interestingly, the percentage of Purα+ irradiated cells in S phase did not change at 6 h and 24 h following irradiation (40.6 and 40.5% respectively), while Purα− cells exited from S phase, (i.e., 42.3% at 6 h and 36.5% at 24 h). These results demonstrate that Purα− cells were able to circumvent the checkpoint despite the presence of unrepaired damaged DNA.
UVC irradiation induces Purα expression in wild-type mouse fibroblasts (MFs).
The experiments performed so far have described a role for Purα in the response to UVC by utilizing Purα− MEFs, which were derived from PURA−/− knockout mice, and comparing them to Purα+ control MEFs, which were made by stably transfecting the Purα− MEFs with an expression plasmid for Purα. Next, we examined the effect of UVC on wild-type MFs where Purα expression is under the control of its own promoter. MFs were synchronized in G1/G0 by serum starvation and treated with and without UVC irradiation. Cells were harvested and western blots performed (). The expression levels from were quantitated by densitometry (). Of note, UVC induced phosphorylation of p53 within 3 hours. At 24 hours, induction of expression of Purα and TFIIH, a multi-subunit transcription/nucleotide excision repair protein, was observed. FACS analysis showed that the UVC-irradiated cells did not progress through the cell cycle (). These data indicate that induction of Purα expression may be a response of the cell to UVC-induced DNA damage.
Purα physically interacts with TFIIH.
Since the previous experiment demonstrated that Purα and TFIIH are induced with similar kinetics by UVC and both proteins are implicated in NER, we next investigated whether they physically interact using an immunoprecipitation/western blot approach to assess binding of Purα with the p89 core subunit of TFIIH, XPB (Xeroderma Pigmentosum B), an ATP dependent DNA helicase. Antibody to TFIIH p89 immunoprecipitated GFP-Purα but not GFP ( and compare lanes 3 and 4). Nonimmune rabbit serum did not immunoprecipitate GFP-Purα (lane 5). We conclude that Purα physically interacts with TFIIH.
Discussion
The single-stranded DNA binding protein Purα is a member of the purine-rich element (PUR) binding protein family and is strongly implicated in the regulation of important steps of nucleic acid processing, including DNA replication, gene transcription, RNA transport and mRNA translation.Citation9 It is also implicated in other important cellular functions, such as growth and proliferation, differentiation and oncogenic transformation.Citation9,Citation17,Citation18 In a previous study, we reported that Purα is involved in conferring resistance to the DNA damage-inducing cancer chemotherapeutic agent cisplatin and Purα deficiency sensitizes cells to treatment with this drug.Citation6 NER is essential for the repair of UV-induced pyrimidine dimers or helix-distorting adducts caused by numerous compounds, including cisplatin.Citation24 Thus we reasoned that Purα may play a role in the cellular response to UV-induced DNA damage. In this study, we utilized fibroblasts derived from wild type mice and MEFs derived from homozygous Purα− knockout mice,Citation22 Purα− and Purα+ control cells, which had Purα re-introduced by ectopic expression. Our data, generated from these cells, clearly indicate that lack of Purα is associated with an increased sensitivity to UVC irradiation. Thus Purα− cells lose viability to a greater extent than Purα+ cells as a function of both UVC dose and time after UVC exposure. Further, recovery of the activity of luciferase enzyme encoded by a reporter plasmid, which had been damaged in vitro by UV irradiation and was introduced into cells by transfection, was impaired in Purα− cells, pointing to deficient DNA repair in the absence of Purα.
We have also previously reported that Purα deficiency hypersensitizes cells to the DNA replication inhibitor hydroxyurea (HU), which inhibits ribonucleotide synthesis and thus depletes the pools of deoxynucleotide triphosphates (dNTP) required for DNA replicationCitation7 and that Purα promotes DNA end joining.Citation6 Taken together, these data suggest that Purα is involved in pathways, which are common for DNA damage response to replication stress, treatment with cisplatin and UV irradiation.
Interestingly, the expression pattern of Purα in response to UV irradiation is similar to the TFIIH p89 subunit, also known as XPB (). Moreover, Purα is physically associated with XPB (), which in eukaryotes is an integral component of the transcription factor TFIIH.Citation25 TFIIH complex is composed of a multi-subunit core and its recruitment is critical to both transcription and NER.Citation26,Citation27 After recognition of lesion-induced distortion of the DNA helix, the TFIIH p89 subunit XPB acts as a helicase and unwinds dsDNA along in a 3′ to 5′ direction in the presence of ATP.Citation28 Endonucleases then incise the damaged DNA strand both 3′ and 5′ to the lesion and resulting DNA gap is refilled by DNA polymerase and rejoined by DNA ligase to complete repair. In this regard, we have found that Purα has an ATP-independent DNA strand-separating activity.Citation29,Citation30 Thus the finding reported here that Purα is associated with TFIIH, a critical factor in NER, taken together with intrinsic qualities of Purα, including its ability to bind to specific DNA sequences and unwind DNA double-strands, suggest the involvement of Purα in nucleoprotein assembly and facilitation of DNA repair at sites of damaged DNA.
Material and Methods
Cell cultures and transfection.
Primary mouse embryo fibroblasts (MEFs) were initially derived from homozygous PURA−/− mouse embryosCitation22 prepared from 13-day pregnant mice using standard techniques. PURA−/−(+Purα) MEFs and PURA−/− (−Purα) MEFs, which were used in these experiments and are abbreviated to Purα+ and Purα− respectively, were established by stably transfecting PURA−/− MEFs with pTRE-Purα containing full length Purα cDNA, which is constitutively expressed, or pcDNA3 empty vector respectively.Citation8 MEFs were cultured in DMEM medium supplemented with 10% FBS and antibiotics. Cells were maintained in a humid incubator at 37°C with 7% CO2. Cells were transfected with plasmids using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Cells were transfected with siRNA using Oligofectamine (InVitrogen) according to the manufacturer's protocol. Primary mouse fibroblasts were isolated from the skin of wild-type Balb/c newborn mice using standard techniques.
Plasmids.
pLEGFPC1 expresses GFP and was from Clontech, (Palo Alto, CA). pLEGFPC1-Purα expresses a GFP-Purα fusion protein and pCMV-Purα (pCDNA6B-Purα) expresses Purα and have been described previously.Citation37 We have also previously described pCMV-luciferase reporter plasmid.Citation6
Treatment of cells with UVC irradiation.
Monolayers of Purα+ and Purα− MEFs were washed with PBS, covered with PBS and irradiated with different doses (0–100 J/m2) of UVC (wavelength = 254 nm, Stratagene UV Stratalinker, Agilent Technologies, Santa Clara, CA) followed by the replacement of the PBS with fresh medium. At designated time points, the cells were harvested and analyzed according to the experimental design.
Protein extracts and western blot analysis.
For whole cell extract preparation, cells were lysed for 30 min on ice in TNN buffer (50 mM Tris-HCl, pH 7.5/150 mM NaCl/0.5% NP-40) containing protease inhibitor cocktail (Sigma, P8340). Cell debris was removed by centrifugation at 14,000 rpm for 10 min at 4°C. Western blot analysis was performed as described.Citation6
Methylthiazoletetrazolium (MTT ) assay.
The MTT assay used the Cell Proliferation Kit I (MTT) according to the manufacturer's protocol (Roche). Purα+ and Purα− cells were irradiated with different doses of UVC as described above. At designated time points, cells were incubated with MTT labeling reagent (final concentration 0.5 mg/ml) for 4 hours at 37°C and the reaction was stopped by the addition of solubilization solution containing 10% SDS in 0.01 M HCl. Viable cells with active mitochondria cleave the tetrazolium ring into a visible dark blue formazan reaction product, which was quantified by spectrophotometry in a microplate reader at 570 nm with a reference wavelength of 650 nm.
Clonogenic assay.
Purα+ and Purα− cells were plated (500 cells/100 mm dish) and treated with or without UVC as described above. Cells were grown for 10–14 days, fixed and stained with methylene blue. Colonies with more than 50 cells were counted and the percentage of colonies formed in treated plates relative to the untreated plates was calculated for each cell type.
Flow cytometric analysis of the cell cycle.
Purα+ and Purα− MEFs were plated and UVC irradiated as described above. Cells were harvested by centrifugation, washed with PBS and fixed in ice-cold ethanol (70% final concentration). After incubation for 24 h at −20°C cells were washed with PBS containing 1% BSA, stained with propidium iodide 10 µg/ml in PBS containing 250 µg/ml RNase A and incubated at 37°C for 30 minutes in the dark before analysis by fluorescence-activated cell sorting (FACS). Cell cycle distribution was analyzed with the GuavaEasy Cyte mini system and using the Guava CytoSoft cell cycle program according to the manufacturer's instructions (Guava Technologies, Hayward, CA). The DNA content determination was measured by the intensity of the PI fluorescence.
Reactivation of UVC-irradiated plasmid.
The ability of cells to repair DNA was determined by transfection with a reporter plasmid with DNA that had been damaged in vitro by UVC irradiation and measuring reconstitution of reporter function. The reporter plasmid pCMV-luciferase in TE pH 7.4 buffer was irradiated with 250–1,000 J/m2 UVC. Purα+ and Purα− cells were transfected with UVC-irradiated or unirradiated control plasmid together with pRL-TK internal control reporter plasmid, which expresses Renilla reniformis luciferase from the Herpes simplex virus thymidine kinase promoter and serves as a control for transfection efficiency. Protein extracts were prepared at various time points after transfection and a luciferase assay was performed using the Promega Dual Luciferase Assay Kit (Promega, Madison, WI) according to the manufacturer's instructions. Quantitation was with a luminometer (Femptomaster FB12, Zylux Corporation). The ratio of Photinus pyralis firefly luciferase activity measured for the irradiated plasmid to that for the control plasmid was calculated for each time point and normalized to Renilla luciferase activity. In one set of experiments, the transfection step also included pCMV-Purα, which expresses Purα or control vector pCMV.
Comet assay.
Comet nuclei formation was analyzed by alkaline Comet assay as we have previously described.Citation31 Serum-starved (48 hr) Purα+ and Purα− cells were treated with or without UVC and analyzed by Comet assay after 24, 48 and 72 hours. The Comet assay was performed under alkaline conditions.Citation32 About 105 cells in 250 µl of PBS were mixed with 750 µl of 1.33% low-melting-point agarose, type VII in PBS (Sigma A-4018). One hundred microliters of cell suspension were spread on a frosted microscope slide that had been precoated with 1% N-agarose, type I-A in H2O (Sigma A-0169). Slides were placed in cold lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Tris [pH 10] and 1% Triton X-100, added freshly before use) for 1 h at 4 °C. Slides were incubated for 40 min in alkaline unwinding buffer (300 mM NaOH and 1 mM EDTA, pH > 13) in the dark at 4°C. Electrophoresis was conducted for 30 min at 25 V (0.72 V/cm) and 300 mA. Slides were washed with distilled H2O 3 × 5 min, air dried and stained for analysis with propidium iodide (PI) (2.5 µg/ml) in sodium citrate pH 8.2 and covered with cover slips. Images of at least 100 cells per sample (50 cells/slide) were evaluated using a fluorescence microscope and Comet 5.0 image analysis software (Kinetic Imaging, Liverpool, UK). Necrotic and apoptotic cells, identified by their microscopic appearance (comets with no heads and nearly all DNA in the tail), were excluded from the analysis.Citation33 The mean of the olive tail moment (OTM) (the product of the tail length and the fraction of the total DNA in the tail) was calculated as a measure of DNA damage.Citation34
Treatment of cells with siRNA targeting Purα.
Cells were treated with either Purα siRNA or non-targeting siRNA using Oligofectamine according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Smartpool Purα siRNA and nontargeting siRNA were obtained from Dharmacon (Lafayette, CO) and were used at a final concentration of 50 nM. In all experiments, western blot analysis for Purα was performed with protein extracts from control cells and cells transfected with Purα−specific siRNA to verify Purα knock-down with anti-α− tubulin antibody used as a loading control.
Antibodies.
The following antibodies were used: Antiphospho-histone H2AX (Ser 139) was from Upstate Biotechnology (Lake Placid, NY). Rabbit polyclonal anti-Purα was as previously described.Citation6 Anti-thymine dimer mouse monoclonal clone KTM53 was from Kamiya Biomedical Company, Seattle, WA. Anti-p53-Ab1 (clone 421) was from CalBiochem (EMD Chemicals, Gibbstown, NJ). Mouse monoclonal anti-α− tubulin (clone B512) was from Sigma (St. Louis, MO). Mouse monoclonal anti-phospho-ATM (Ser 1981), rabbit polyclonal anti-phospho-ATR (Ser 428) and rabbit polyclonal anti-phospho-p53 (Ser 15) were purchased from Cell Signaling (Danvers MA). Rabbit polyclonal anti-TFIIH p89 (S-19) and mouse monoclonal anti-PCNA (PC-10) antibodies were from Santa Cruz Biotechnology, Inc., (Santa Cruz, CA). Rabbit polyclonal anti-RPA 32 was from GeneTex (Irvine, CA). Rabbit polyclonal antibody to GFP (Living Colors) was from Clontech (Palo Alto, CA).
Detection of cyclobutane pyrimidine dimers (CPDs) by FACS analysis.
Flow cytometry-based detection of CPDs in UVC-irradiated cells was performed as describedCitation35 with minor modifications. Briefly, Purα+ and Purα− cells were synchronized in G0/G1 by 48 h serum deprivation, washed with PBS and while in PBS were irradiated with 10 J/m2 UVC. After 24 h or at time zero, cells were washed with PBS/50 mM EDTA solution, trypsinized and resuspended in 1 ml of PBS/50 mM EDTA. Cells were fixed with ice-cold ethanol (70%, final concentration), washed with PBS/50 mM EDTA, gently resuspended in 0.5% Triton X-100/2 N HCl and incubated for 20 min at 22°C. Cells were washed with 0.1 M Na2B4O7 (pH 9.0) and then with PBS and resuspended in 300 µl of RNase (100 µg/ml in PBS) for 1 h at 37°C. After washing with PBS-TB (PBS/1% BSA/0.25% Tween 20), cells were incubated with primary anti-Thymine dimer antibody for 1 hour at room temperature, washed with PBS-TB and resuspended in 300 µl of PBS-TB containing fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse secondary antibody (Sigma) for 45 minutes at room temperature in the dark. As a negative control, samples were left without anti-Thymine dimer antibody but FITC-conjugated secondary antibody alone was applied. After washing cells with PBS-TB, cells were stained with propidium iodide (5 µg/ml) in PBS. Repair kinetics were monitored using the GuavaEasy Cyte mini system and using the Guava CytoSoft cell cycle program according to the manufacturer's instructions (Guava Technologies, Hayward, CA). The DNA content determination was measured by the intensity of the PI fluorescence.
Detection of cyclobutane pyrimidine dimers (CPDs) by slot blot analysis.
CPDs induced by UV-treatment were detected as described by Guo et al.Citation36 with minor modifications. Briefly, Purα+ and Purα− MEFs were synchronized in G1/G0 by serum deprivation for 48 hours and treated with 10 J/m2 of UVC at the same time as release from serum starvation. Control cultures were left untreated. Cells were harvested at 5 min, 24 h and 48 h following UV treatment and genomic DNA was isolated using the Archive Pure Cell/Tissue Kit (5 Prime, Inc., Gaithersburg, MD). One hundred nanograms of DNA were spotted on a supported Optitran nitrocellulose membranes (Whatman, Piscataway, NJ) using slot-blot Bio-Dot SF transfer apparatus (Bio-Rad, Hercules, CA). CPDs formed as a result of UV-induced DNA damage were detected by immunoblotting, using monoclonal antibody against CPD (1:1,000). The densities of the bands were quantified by densitometry and the graphs were plotted after normalization to the amount of loaded DNA.
Effect of UV on cells synchronized by thymidine block.
Purα+ and Purα− cells were plated at a density of 3 × 105 cells/100 mm dish and synchronized in S phase by double thymidine block (G1/S block, 2 mM thymidine). After 48 hours, cells were washed in PBS and irradiated with 50 J/m2 of UVC. After 6 and 24 h, cells were harvested and samples prepared for FACS and western blot analysis.
Co-immunoprecipitation assay.
Purα− MEFs were transfected with pLEGFPC1 or pLEGFPC1-Purα.Citation37 After 48 hrs, cells were harvested and immunoprecipitation/western blots performed as previously describedCitation38 using antibody to p89 of TFIIH for precipitation and antibody to GFP for the western blot. Precipitation with nonimmune rabbit serum was the negative control. One tenth of each input was loaded as positive control.
Figures and Tables
Figure 1 Cell viability and clonogenicity of Purα-positive (Purα+) and Purα-negative (Purα−) cells in response to UVC treatment. (A) Purα+ and Purα− MEFs were treated with different doses of UVC and assayed for viability after 96 h by MTT assay. Values obtained for each UVC dose were normalized relative to UVC-untreated Purα+ cells. ■-Purα+; □-Purα+. (B) Purα+ and Purα− MEFs were treated with 10 J/m2 UVC and MTT assays performed at the time points indicated. Values were normalized relative to UVC-untreated cells at 24 h. ■-Purα+ untreated; ▴-Purα+ treated with 10 J/m2 UVC; □-Purα− untreated;. ▵-Purα− treated with 10 J/m2. (C) Purα+ and Purα− MEFs were transfected with non-target siRNA or siRNA specific for Purα at 24 h before UVC treatment (10 J/m2) and MTT assays were performed after 96 h. Values were normalized relative to UVC-untreated cells at 24 h. ■-Purα+ + NT siRNA-UVC; □-Purα+ + Purα si RNA-UVC; ▴-Purα+ + NT siRNA + UVC; ▵-Purα+ + Purα si RNA + UVC. (D) Purα+ and Purα− MEFs were transfected with pCMV plasmid or pCMV-Purα expression plasmid, treated with different doses of UVC and assayed for viability after 96 h by MTT assay. Values were normalized to UVC-untreated MEF Purα− (+pCMV) cells. ■-Purα+ + pCMV-Purα; □-Purα+ + pCMV. (E) Western blot for Purα for the cells used in (A and B) with α-tubulin as a loading control. (F) Western blot for Purα for the cells used in (C) with α-tubulin as a loading control. (G) Western blot for Purα for the cells used in (D) with α-tubulin as a loading control. (H) Purα+ and Purα− MEFs were treated with UVC as indicated and plated. After two weeks, colonies were counted and values normalized relative to unirradiated controls. The experiments were repeated five times. *p < 0.03.
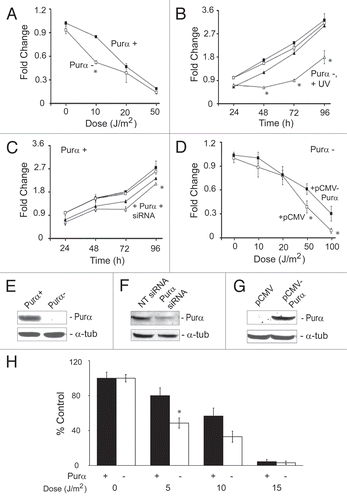
Figure 2 Reactivation of transfected UVC-irradiated reporter plasmid in Purα+ and Purα− cells. Luciferase reporter plasmid was treated in vitro with and without UVC as described in Materials and Methods and then introduced into Purα+ and Purα− cells by transfection. (A) Dose-response for restoration of luciferase activity. (B) In this experiment, expression vectors pCMV-Purα and pCMV were included in the transfections together with the irradiated plasmid. The experiments were repeated at least three times. *p < 0.03.
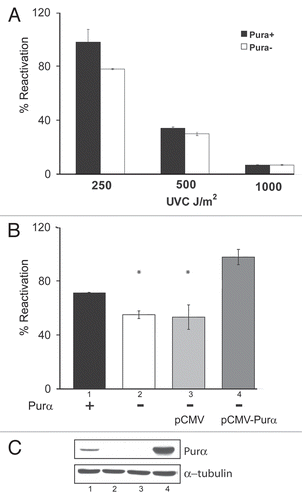
Figure 3 Assays of cyclobutane pyrimidine dimers (CPDs) in UV-irradiated Purα+ and Purα− cells. (A) Purα+ and Purα− cells were synchronized by 48 h serum deprivation and treated with 10 J/m2 of UVC. After 24 h or at time zero, cells were fixed with 70% ethanol and labeled for CPDs with anti-thymine antibody and FITC-conjugated secondary antibody. After adding propidium iodide, G1/G0 cell populations were analyzed by flow cytometry for CPDs. Histograms show fractions of CPD-positive cells in green in the analyzed samples. (B) Purα+ and Purα− MEFs were synchronized in G1/G0, treated with UVC, DNA isolated and analyzed by slot immunoblotting using monoclonal antibody against CPD as described in Materials and Methods. The densities of the bands were quantified by densitometry and the graphs were plotted after normalization to the amount of loaded DNA. The experiments were repeated at least three times. *p < 0.05.
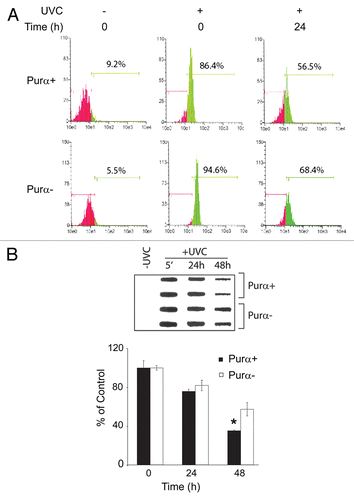
Figure 4 DNA fragmentation measured by the Comet assay in UV-irradiated Purα+ and Purα− cells. (A) Purα+ and Purα− cells were synchronized by 48 h of serum deprivation, treated with 50 J/m2 of UVC irradiation, subject to in situ electrophoresis and the tail of fragmented DNA measured (Olive Tail Moment) as described in Materials and Methods (Comet assay). The OTM was scored randomly from 100 propidium iodide labeled cells, for each condition. The plot shows a percentage of the Comet positive nuclei (OTM>13), apoptotic and necrotic cells were not scored. (B) Representative photomicrograph of cells with low and high OTM. The experiments were repeated at least three times.
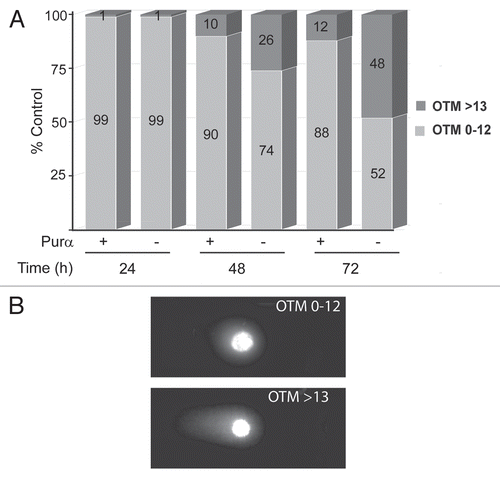
Figure 5 Effect of UVC on DNA damage signaling pathways in Purα+ and Purα− cells. Purα+ and Purα− cells were synchronized by double thymidine block (G1/S block, 2 mM thymidine) and treated with 50 J/m2 of UVC. After 6 and 24 h, whole cell protein extracts were analyzed by western blot.
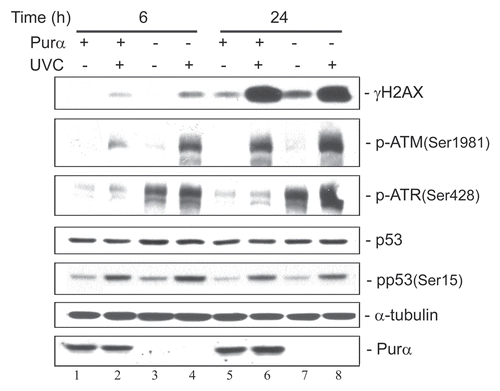
Figure 6 Effect of UVC irradiation on synchronized wild-type Balb/c mouse fibroblasts. Wild-type Balb/c mouse fibroblasts were synchronized by 72 hours of serum starvation and then treated with and without UVC (20 J/m2) at time zero. (A) Cells were harvested and subject to western blot. (B) The western blots in (A) were quantitated by densitometry. (C) Cells from the same samples were harvested and analyzed by FACS for the cell cycle.
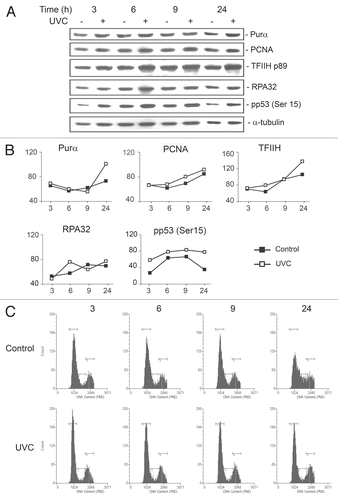
Figure 7 Co-immunoprecipitation of GFP-Purα with TFIIH. Purα− MEFs were transfected with expression plasmid for GFP or GFP-Purα and whole cell extracts prepared. Immuno-precipitation was performed with antibody for TFIIH/p89 (lanes 3 and 4) or nonimmune rabbit serum (NRS-lane 5) and the immune complex analyzed by western blot using antibody to GFP. Lanes 1 and 2 shows 1/10 of the immunoprecipitation input from the GFP- and GFP-Purα−transfected whole cell extracts respectively.
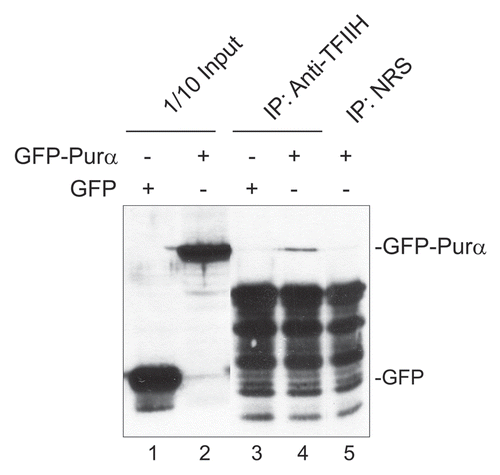
Table 1 Cell cycle profiles of Purα+ and Purα− cells with or without UVC irradiation
Acknowledgements
We thank past and present members of the Center for Neurovirology for their insightful discussion and sharing of ideas and reagents especially Dr. Nune Darbinian. We also wish to thank C. Schriver for editorial assistance. This work was supported by grants awarded by the NIH to M.K.W., E.J. and K.K.
References
- Batista LF, Kaina B, Meneghini R, Menck CF. How DNA lesions are turned into powerful killing structures: insights from UV-induced apoptosis. Mutat Res 2009; 681:197 - 208
- Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol B 2001; 63:88 - 102
- Ford JM. Regulation of DNA damage recognition and nucleotide excision repair: another role for p53. Mutat Res 2005; 577:195 - 202
- Mitchell DL, Jen J, Cleaver JE. Relative induction of cyclobutane dimers and cytosine photohydrates in DNA irradiated in vitro and in vivo with ultraviolet-C and ultraviolet-B light. Photochem Photobio 1991; 54:741 - 746
- Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 2001; 8:213 - 224
- Kaminski R, Darbinyan A, Merabova N, Deshmane SL, White MK, Khalili K. Protective role of Puralpha to cisplatin. Cancer Biol Ther 2008; 7:1926 - 1935
- Wang H, Wang M, Reiss K, Darbinian-Sarkissian N, Johnson EM, Iliakis G, et al. Evidence for the involvement of Puralpha in response to DNA replication stress. Cancer Biol Ther 2007; 6:596 - 602
- Wang H, White MK, Kaminski R, Darbinian N, Johnson EM, Amini S, et al. Role of Purα in the modulation of homologous recombination-directed DNA repair by HIV-1 Tat. Anticancer Res 2008; 28:1441 - 1448
- White MK, Johnson EM, Khalili K. Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle 2009; 8:1 - 7
- Rosell R, Taron M, Barnadas A, Scagliotti G, Sarries C, Roig B. Nucleotide excision repair pathways involved in Cisplatin resistance in non-small-cell lung cancer. Cancer Control 2003; 10:297 - 305
- Haas S, Gordon J, Khalili K. A developmentally regulated DNA-binding protein from mouse brain stimulates myelin basic protein gene expression. Mol Cell Biol 1993; 13:3103 - 3112
- Haas S, Thatikunta P, Steplewski A, Johnson EM, Khalili K, Amini S. A 39 kD DNA-binding protein from mouse brain stimulates transcription of myelin basic protein gene in oligodendrocytic cells. J Cell Biol 1995; 130:1171 - 1179
- Bergemann AD, Johnson EM. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol 1992; 12:1257 - 1265
- Bergemann AD, Ma ZW, Johnson EM. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded DNA-binding properties of the encoded protein. Mol Cel Biol 1992; 12:5673 - 5682
- Ma ZW, Bergemann AD, Johnson EM. Conservation in human and mouse Pur alpha of a motif common to several proteins involved in initiation of DNA replication. Gene 1994; 149:311 - 314
- Liu H, Johnson EM. Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res 2002; 30:2417 - 2426
- Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res 2000; 28:3197 - 3205
- Johnson EM. The Pur protein family: clues to function from recent studies on cancer and AIDS. Anticancer Res 2003; 23:2093 - 2100
- Kaminski R, Darbinian N, Sawaya BE, Slonina D, Amini S, Johnson EM, et al. Puralpha as a cellular co-factor of Rev/RRE-mediated expression of HIV-1 intron-containing mRNA. J Cell Biochem 2008; 103:1231 - 1245
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 2004; 43:513 - 525
- Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, et al. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res 2006; 83:929 - 943
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, et al. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol 2003; 23:6857 - 6875
- Mitchell DL, Nairn RS. The biology of the (6-4) photoproduct. Photochem Photobiol 1989; 49:805 - 819
- Zamble DB, Lippard SJ. Cisplatin and DNA repair in cancer chemotherapy. Trends Biochem Sci 1995; 20:435 - 439
- Oksenych V, de Jesus BB, Zhovmer A, Egly JM, Coin F. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J 2009; 28:2971 - 2980
- Roy R, Schaeffer L, Humbert S, Vermeulen W, Weeda G, Egly JM. The DNA-dependent ATPase activity associated with the class II basic transcription factor BTF2/TFIIH. J Biol Chem 1994; 269:9826 - 9832
- Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, et al. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol Cell 2002; 10:1163 - 1174
- Hwang JR, Moncollin V, Vermeulen W, Seroz T, van Vuuren H, Hoeijmakers JH, Egly JM. A 3′→5′ XPB helicase defect in repair/transcription factor TFIIH of xeroderma pigmentosum group B affects both DNA repair and transcription. J Biol Chem 1996; 271:15898 - 15904
- Darbinian N, Gallia GL, Khalili K. Helix-destabilizing properties of the human single-stranded DNA- and RNA-binding protein Purα. J Cell Biochem 2001; 80:589 - 595
- Wortman MJ, Johnson EM, Bergemann AD. Mechanism of DNA binding and localized strand separation by Purα and comparison with Pur family member, Purβ. Biochem Biophys Acta 2005; 1743:64 - 78
- Merabova N, Kaniowska D, Kaminski R, Deshmane SL, White MK, Amini S, et al. JC virus agnoprotein inhibits in vitro differentiation of oligodendrocytes and promotes apoptosis. J Virol 2008; 82:1558 - 1569
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 1988; 175:184 - 191
- Speit GH, Hartmann A. The comet assay (single-cell gel test). A sensitive genotoxicity test for the detection of DNA damage and repair. Methods Mol Biol 1999; 113:203 - 212
- Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat Res 1990; 122:86 - 94
- Rouget R, Auclair Y, Loignon M, Affar el B, Drobetsky EA. A sensitive flow cytometry-based nucleotide excision repair assay unexpectedly reveals that mitogen-activated protein kinase signaling does not regulate the removal of UV-induced DNA damage in human cells. J Biol Chem 2008; 283:5533 - 5541
- Guo R, Chen J, Zhu F, Biswas AK, Berton TR, Mitchell DL, et al. E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J Biol Chem 2010; 285:19308 - 19315
- Darbinian N, White MK, Khalili K. Regulation of the Pur-alpha promoter by E2F-1. J Cell Biochem 2006; 99:1052 - 1063
- White MK, Skowronska A, Gordon J, Del Valle L, Deshmane SL, Giordano A, et al. Analysis of a mutant p53 protein arising in a medulloblastoma from a mouse transgenic for the JC virus early region. Anticancer Res 2006; 26:4079 - 4092