Abstract
Sister chromatid pairing reactions, termed cohesion establishment, occur during S-phase and appear to be regulated by Replication Factor C (RFC) complexes. For instance, RFCs that contain Ctf18p exhibit pro-establishment activities while those that contain Elg1p exhibit anti-establishment activities. It remains unknown whether Ctf18p-RFC and Elg1p-RFC functions are simply opposing or instead reveal complicated and non-parallel regulatory mechanisms. To better understand the nature of these novel pathways, we analyzed the small RFC subunit Rfc5p that is common to both Ctf18p-RFC and Elg1p-RFC. Despite this commonality, the data show that diminished Rfc5p function rescues ctf7/eco1 mutant cell phenotypes, revealing that Rfc5p promotes anti-establishment activities. This rescue is specific to establishment pathways in that rfc5-1 greatly accentuates growth defects when expressed in scc2 (deposition), mcd1/scc1 or smc3 (cohesion maintenance) mutated cells. Our results reveal for the first time a role for small RFC subunits in directing RFC complex functions - in this case towards anti-establishment pathways. We further report that Pds5p exhibits both establishment and anti-establishment functions in cohesion. This duality suggests that categorizations of establishment and anti-establishment activities require further examination.
Introduction
A typical cell cycle invariably includes a DNA replication step to produce two exact copies of each chromosome. The replication factor C complex (RFC) is a five subunit complex consisting of one large subunit (Rfc1p) accompanied by four small subunits (Rfc2p-Rfc5p) and is required for DNA replication. RFC complexes hydrolyze ATP to open and then load PCNA or PCNA-like sliding clamps onto primer-template regions of DNA. In turn, sliding clamps maintain polymerase association with DNA and thus promote processive DNA replication.Citation1 All five of the canonical RFC subunits (Rfc1p-Rfc5p) and PCNA are essential for DNA replication during S phase. In addition to Rfc1p, there are three alternative large subunits (Ctf18p, Elg1p and Rad24p) that exhibit extensive sequence homology to Rfc1p and associate individually with each of the four small subunits Rfc2p-Rfc5p. Ctf18p-RFC both binds PCNA and exhibits PCNA loading/unloading activities.Citation2,Citation3 Elg1p-RFC also binds PCNA, but the extent to which this complex promotes sliding clamp association with DNA remains unknown.Citation4–Citation6 Rad24p-RFC is unique in associating with the 9-1-1 sliding clamp complex composed of Rad17p, Mec3p and Ddc1p.Citation7–Citation9 More recent evidence reveals that Rad24p-RFC can also bind and elicit PCNA unloading.Citation10 Notably, all four RFC complexes and both PCNA and Rad17p, Mec3p, Ddc1p sliding clamps function in DNA repair pathways.Citation11
In order for the products of DNA replication to be segregated properly in mitosis, a multi-step cohesion pathway is required earlier during the cell cycle. During S phase for instance, the products of chromosome replication, termed sister chromatids, become decorated with cohesin complexes that ultimately serve as a molecular glue that tethers the sisters together. Cohesins contain both structural components (Smc1p, Smc3p, Mcd1p/Scc1p and Scc3p/Irr1p) and accessory factors (Pds5p and Rad61p).Citation12,Citation13 Before chromatid-associated cohesins can participate in sister chromatid pairing, however, they must be modified by Ctf7p/Eco1p—an aceytltransferase that targets Smc3p specifically during S phase.Citation14–Citation16 In the absence of this establishment step, sister chromatids remain cohesin-decorated but unpaired, resulting in massive chromosome missegregation and cell death.Citation17,Citation18 Several models of the cohesin structures that maintain sister chromatid pairing remain actively debated.Citation19
A series of findings now reveal that RFC complexes also exhibit critical roles in sister chromatid pairing reactions.Citation20–Citation24 Early studies revealed that Ctf18p-RFC promotes sister chromatid pairing reactions and is capable of binding the establishment factor Ctf7p/Eco1p in vitro.Citation17,Citation20–Citation22 More recent findings reveal that mutation in Elg1p-RFC also elicits sister chromatid pairing defects and that Elg1p binds Ctf7p/Eco1p in vitro.Citation23,Citation24 Importantly, however, loss of ELG1 rescues ctf7/eco1 mutant cell defects, in stark contrast to the lethality resulting from loss of CTF18 in ctf7/eco1 mutant cells. A simple interpretation of these findings is that Ctf18p-RFC and Elg1p-RFC perform opposing pro- versus anti-establishment activities, respectively, and that the activities of these RFC complexes are directed by the associations of the unique large RFC subunit.Citation25 Large RFC subunits are commonly considered as directing alternate RFC complex functions in DNA replication and DNA repair pathways.
Despite the vast majority of studies that focus on the role of large RFC subunits in dictating RFC complex functions in DNA replication, repair and sister chromatid cohesion, there is limited evidence that small subunit assemblies are sufficient for RFC activity such as clamp unloading.Citation26 Here, we report on a novel and critical role for the small RFC subunit Rfc5p in directing anti-establishment activities of alternate RFC complexes and discuss new models regarding the regulatory mechanisms that drive RFC functions in vivo.
Results
Rfc5p directs RFC-dependent sister chromatid pairing regulation.
Ctf18p-RFC is designated a pro-establishment factor in part based on observations that CTF18 deletion is lethal when combined with ctf7eco1 mutations.Citation17 Conversely, Elg1p-RFC is categorized as an anti-establishment factor since ELG1 deletion rescues ctf7eco1 mutant cell growth and cohesion defects.Citation23,Citation24 RFC small subunits must also contribute to cohesion-dependant activities given that mutations in RFC4 or RFC5 produce cohesion defects.Citation21,Citation22 Despite these findings, analyses that test for the role of small RFC subunits in directing RFC complex function toward either pro- or anti-cohesion establishment are absent from the literature. We reasoned that diminishing Rfc5p function in ctf7eco1 mutant cells might produce a balanced reduction of Ctf18p-RFC and Elg1p-RFC and would thus be transparent to cohesion pathways. Alternatively, if small RFC subunits contribute in a biased fashion to cohesion regulation, then diminished Rfc5p function might elicit defects specifically in either establishment or anti-establishment activities. This difference is resolvable by testing for lethality or rescue of ctf7eco1 mutant cell phenotypes, respectively.Citation19 To differentiate between these possibilities, ctf7eco1-1 mutant cells were crossed to rfc5-1 cells, sporulated and the resulting diploids dissected. We obtained the predicted number of single mutant ctf7eco1 and rfc5 cells and also of double mutant ctf7eco1 rfc5 cells. Thus, the combination of rfc5 and ctf7eco1 does not phenocopy the lethality of ctf7 ctf18 double mutant cells. We then tested whether rfc5-1 exacerbates or rescues conditional growth defects of ctf7eco1 mutant cells. Serial dilutions of log phase wild-type, single and double mutant cell cultures were plated onto rich media and maintained at either 23°C, 27°C or 37°C. As expected, both ctf7eco1 and rfc5 single mutant cells are inviable at 37°C. ctf7eco1 mutant cells exhibit extreme temperature sensitivity in that these cultures are inviable even at 27°C (). Surprisingly, the addition of the rfc5 mutation rescues ctf7eco1 conditional lethality such that ctf7 rfc5 double mutant cells exhibit growth at temperatures otherwise lethal for ctf7eco1 mutant cells (). We note that the rfc5-dependent rescue of ctf7eco1 mutant cell defects appears identical to that produced by deletion of elg1 in ctf7eco1 mutant cells.Citation23 Moreover, the additional deletion of ELG1 from rfc5-1 ctf7eco1-1 mutant cells did not further enhance growth, suggesting that Elg1p and Rfc5p operate through a common pathway (data not shown). In combination, these studies suggest that the small Rfc5p subunit is a critical regulator of RFC complex function during sister chromatid pairing reactions.
Rfc5 supports cohesin maintenance pathways.
While deletion of ELG1 rescues ctf7/eco1 establishment defects, it greatly exacerbates the conditional growth observed in smc1, smc3, mcd1/scc1 and scc3/irr1 mutant cells (cohesion maintenance) and also in scc2 and scc4 (cohesin deposition) mutant cells.Citation23,Citation24 Thus, it becomes important to test whether Rfc5p similarly supports cohesin maintenance pathways. rfc5-1 and mcd1-1/scc1 single mutant strains were crossed, the diploids sporulated and dissected to obtain wild-type, single and double mutant cells. We obtained roughly the expected number of rfc5-1 mcd1-1/scc1 double mutant cells, along with single mutant and wild-type strains. Serial dilutions of log phase strains were plated onto rich medium and grown at a range of temperatures. As expected, rfc5-1 and mcd1-1/scc1 single mutant cells exhibited robust growth at temperatures up to 30°C but were inviable when maintained at 37°C. In contrast, rfc5-1 mcd1-1/scc1 double mutant cells are largely inviable at 30°C (). A single revertant rfc5-1 mcd1-1/scc1 double mutant spore is currently under investigation. We repeated our analysis but this time using a different cohesin complex mutant strain, smc3-5 cells. Serial dilutions of log phase strains obtained from sporulated diploids of rfc5-1 crossed to smc3-5 revealed that diminished Rfc5 adversely affects smc3-5 mutant cell growth such that the double mutant cells are largely inviable at 32° (). We decided to include elg1 deletion strains in the above crosses to aide in the comparison of elg1 and rfc5 effects. The results show that, similar to rfc5-1, deletion of elg1 from smc3-5 mutant cells greatly exacerbates the conditional growth phenotype. Even at the permissive temperature of 23°C, elg1 smc3-5 double mutant cells exhibit diminished growth. This adverse effect was less detectable in rfc5-1 smc3-5 double mutant cells (). These findings reveal the participation of Rfc5p in supporting cohesion maintenance pathways.
Rfc5 supports Scc2-dependent cohesin deposition pathways.
The conversion of cohesins to a paired state (establishment) occurs only after cohesin deposition onto chromatin. Indeed, numerous studies document that deposition and establishment are temporally and genetically separable.Citation17,Citation18,Citation27,Citation28 We decided to exploit these differences to test directly whether the Rfc5p small subunit supports sister chromatid pairing accruing through cohesin deposition pathways. rfc5-1 and scc2-4 single mutant cells were mated, the diploids sporulated and wild-type, single and double mutant strains isolated. We included elg1 deletion strains in the matings for comparison. Serial dilutions of the resulting log phase cultures confirmed that both rfc5-1 and scc2-4 single mutant strains are inviable at 37°C but exhibit fairly robust growth at 30°C. However, the results show that both scc2-4 elg1 and scc2-4 rfc5 double mutant strains exhibit greatly exacerbated growth defects even at 30°C, with elg1 deletion producing the more significant challenge to scc2-4 mutant cell viability (). Thus, RFC complex functions are crucial very early in cohesion pathways and specifically during S-phase-dependent cohesin deposition and possibly as early as late G1 phase.
Pds5 exhibits both pro- and anti-establishment activities.
Pds5p binds cohesins, and Pds5p activity is essential for maintaining cohesion until anaphase onset. In addition, several lines of evidence document that Pds5p also regulates cohesion by exhibiting anti-establishment activities.Citation29,Citation30 Despite the importance of Pds5p in cohesion, the mechanism through which it performs anti-establishment activity remains unclear. We decided to capitalize on our findings that either RFC5 or ELG1 deletion rescues establishment mutant cell phenotypes but aggravates cohesin maintenance and deposition of mutant cell phenotypes. To identify in which pathway Pds5p may function, elg1 and rfc5-1 single mutant strains were individually crossed to pds5-1 single mutant cells and the resulting diploids sporulated. Serial dilutions of the resulting wild type, single and double mutant cells were plated onto rich medium and colony growth challenged at a range of temperatures. Diminished Rfc5p function neither rescues nor aggravates pds5-1 mutant cell growth defects within the range of temperatures tested (). Surprisingly, however, the results show that deletion of ELG1 suppresses pds5-1 mutant cell growth defects even at 37° (). This current study, in combination with prior studies, reveal that elg1 deletion suppresses defects that occur through establishment mechanisms.Citation23,Citation24 These findings suggest that Pds5p exhibits an establishment activity in addition to a documented role as an anti-establishment factor.
Discussion
Historically, the role of Rfc1p-RFC complex has been relatively unidimensional: to load/unload PCNA sliding clamp at primer template sites to ensure processive DNA replication. With the identification of multiple large RFC subunits (Ctf18p, Rad24p and Elg1p) unique to alternate RFC complexes, this role has expanded in numerous and important ways. First, the role of RFCs was extended to include DNA damage response and checkpoint function. Second, at least one RFC was found to load/unload the 9-1-1 sliding clamp (Rad17p, Mec3p, Ddc1p) instead of PCNA.Citation11,Citation31,Citation32 Third, characterization of RFC complexes that incorporate Ctf18p revealed a pro-establishment activity crucial for sister chromatid pairing.Citation17,Citation20,Citation21 More recently, the role in cohesion expanded even further to include an Elg1p-RFC-dependent anti-establishment activity.Citation23,Citation24 Up to this point, the various roles for RFC complexes were attributed to the identity of the unique large RFC subunit. Here, we challenge this notion and show for the first time that small RFC subunits such as Rfc5p play a critical role in determining RFC complex function. While our data is specific for sister chromatid cohesion reactions, we speculate that small RFC subunit-directed activities may be wide-spread and predict similar findings in regulating DNA replication and various DNA repair pathways.
Our finding that Rfc5p is capable of biasing entire RFC complex function with respect to cohesion suggests several mechanisms (). For instance, small subunits may contribute directly to biasing RFC complex function (). Possible scenarios include that small subunits such as Rfc5p provide a cooperative binding site for cohesins or cohesin deposition-modified chromatin crucial for directing/recruiting RFC complex function. Small RFC subunits may also act directly in cohesion as sensors or signal transducers, consistent with Rfc5p function in checkpoint pathways.Citation33–Citation35 Alternatively, Rfc5p may indirectly regulate RFCs by differentially regulating large subunit association in response to environmental conditions. In the current study, this particular allele of Rfc5p may inactivate the Elg1p-RFC anti-establishment complex but not the Ctf18p-RFC pro-establishment complex via differential binding (). In either case, these scenarios highlight novel mechanisms through which small subunits may regulate RFC complex functions. While less likely, we further speculate that large RFC subunits may associate with some but not all small RFC subunits in differentiating between pro- and anti-establishment activities. In support of this latter model is evidence that Ctf18p binds Dcc1p and Ctf8p during cohesion establishment and that small RFC subunits appear to perform separable reactions in cohesion and DNA repair ().Citation20–Citation22,Citation30,Citation36 Lastly, RFC small subunit assemblies devoid of large subunits are competent to drive sliding clamp loading, and this suggests that small subunits may be capable of forming a unique anti-establishment clamp loader/unloader complex.Citation26 While the current study focuses on the small Rfc5p subunit in cohesion, these findings are likely to have broad implications regarding subunit contributions to DNA replication and directing alternate RFC complexes between various DNA repair pathways.
The mechanism through which Elg1p-RFC opposes Ctf7p/Eco1p activity is poorly understood. Our finding that deletion of ELG1 suppresses the growth defects of pds5 mutant cells suggest that Pds5p promotes establishment in coordination with Ctf7p/Eco1p activity. In support of this model are early studies that document an essential role for Pds5p in maintaining sister chromatid pairing.Citation37–Citation39 A pro-establishment role is further supported by findings that Ctf7p/Eco1p physically binds Pds5p in vitro and that mutations of either are highly sensitive to changes in dosage of the other.Citation40 The combination of these findings is in stark contrast to the characterization of Pds5 as an antiestablishment factor.Citation29,Citation41,Citation42 The current study unambiguously points to the duality of pro- and anti-establishment regulation of sister chromatid pairing by Pds5p. We speculate that Pds5p functions in a multistep pathway in which its binding partners, modifications or other regulatory mechanisms switch Pds5p between pro- and anti-establishment activities. It is thus important that the combined impact of these studies direct new investigations aimed at elucidating the bi-functionality of factors involved in sister chromatid pairing reactions.
In several, but not all, instances we have uncovered evidence separating Elg1p function from that of Rfc5p. First, elg1 smc3-5 double mutant cells exhibit diminished growth beyond that observed for rfc5-1 smc3-5 double mutants. Second, elg1 deletion produced more significant challenges to scc2-4 mutant cell viability than the rfc5-1 hypomorph. Third and most importantly, deletion of ELG1 (but not expression of rfc5-1 hypomorph) dramatically suppressed pds5-1 mutant cell growth defects even at elevated temperatures. Despite these differences, both elg1 and rfc5-1 suppress ctf7/eco1 mutant cell phenotypes to an identical extent. These findings obviate concerns that rfc5-1 is simply mimicking elg1 deletion. More importantly, they reveal the underappreciated complexity of RFC complex function in cohesion pathways—and likely also those in DNA replication and repair.
Materials and Methods
Yeast strains and media.
All strains used in this study are W303 background unless otherwise noted (). Diploid strains were sporulated in 0.3% potassium acetate and tetrads dissected on YPD media.Citation43 The genotypes of the resultant spores were analyzed for each wild-type, single and double mutant spore recovered. Phenotypic analyses of isolated spores was performed as previously described.Citation23 Briefly, log phase cultures grown in rich liquid YPD medium were normalized and used to generate 10-fold serial dilutions of each strain. Each dilution series was plated on rich YPD agar plates and grown at a range of temperatures as indicated in each figure.
Figures and Tables
Figure 1 rfc5-1 mutation suppresses ctf7eco1-1 mutant cell conditional growth. Growth of 10-fold serial dilutions of wildtype, ctf7eco1-1 and rfc5-1 single mutant strains compared to that of ctf7eco1-1 rfc5-1 double mutant strains (three independent isolates shown). Colony growth shown for cells on rich medium plates maintained at 23°C, 27°C and 37°C.
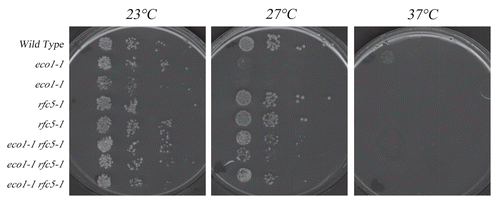
Figure 2 rfc5-1 mutation exacerbates mcd1-1 mutant cell conditional growth. Growth of 10-fold dilutions of wild type, rfc5-1 and mcd1-1 single mutant strains and rfc5-1 mcd1-1 double mutant strains (two independent isolates shown). Colony growth shown on rich medium plates maintained at 23°C, 30°C and 37°C.
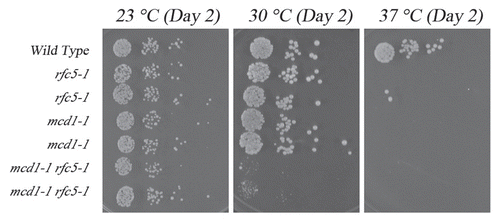
Figure 3 Growth analyses of smc3-5 cohesion-defective strains in the presence of additional rfc5-1 or elg1 mutations. Top row: rfc5-1 mutation exacerbates smc3-5 mutant cell conditional growth. Growth of 10-fold dilutions of wild-type, rfc5-1 and smc3-5 single mutant strains and rfc5-1 smc3-5 double mutant strains (three independent isolates shown). Colony growth shown on rich medium plates maintained at 23°C, 30°C, 32°C and 37°C. Minor E. coli colonies also present in wild-type patch. Bottom row: elg1 deletion exacerbates smc3-5 mutant cell conditional growth. Growth of 10-fold dilutions of wild-type, elg1and smc3-5 single mutant strains and elg1 smc3-5 double mutant strains (three independent isolate shown). Colony growth shown on rich medium plates maintained at 23°C, 30°C and 37°C.
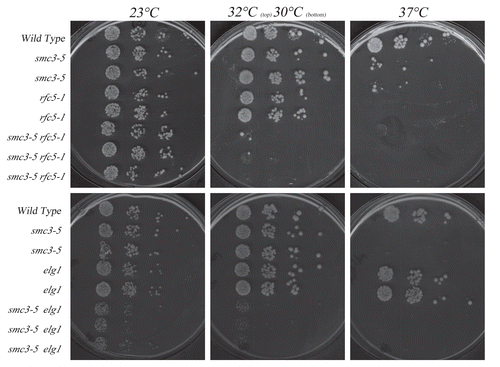
Figure 4 Growth analyses of scc2 cohesion-defective strains in the presence of additional rfc5-1 or elg1 mutations. Top row: rfc5-1 mutation exacerbates scc2-4 mutant cell conditional growth. Growth of 10-fold dilutions of wild type, rfc5-1 and scc2-4 single mutant strains and rfc5-1 scc2-4 double mutant strains (three independent isolates shown). Colony growth shown on rich medium plates maintained at 23°C, 30°C and 37°C. Bottom row: elg1 deletion exacerbates scc2-4 mutant cell conditional growth. Growth of 10-fold dilutions of wild type, elg1 and scc2-4 single mutant strains and elg1 scc2-4 double mutant strains (three independent isolates shown). Colony growth shown on rich medium plates maintained at 23°C, 30°C and 37°C.
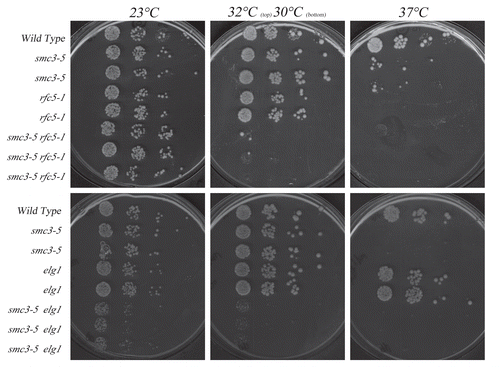
Figure 5 Growth analyses of pds5-1 cohesion-defective strains in the presence of additional rfc5-1 or elg1 mutations. Top row: rfc5-1 mutation neither suppresses nor exacerbates pds5-1 mutant cell conditional growth. Growth of 10-fold serial dilutions of wild-type, pds5-1 and rfc5-1 single mutant strains and pds5-1 rfc5-1 double mutant strains (three independent isolates shown). Colony growth shown for cells on rich medium plates maintained at 23°C, 27°C and 37°C. Bottom row: elg1 deletion suppresses pds5-1 mutant cell conditional growth. Growth of 10-fold serial dilutions of wild-type, pds5-1 and elg1 single mutant strains compared to pds5-1 elg1 double mutant strains (three independent isolates shown). Colony growth shown for cells on rich medium plates maintained at 23°C, 27°C and 37°C.
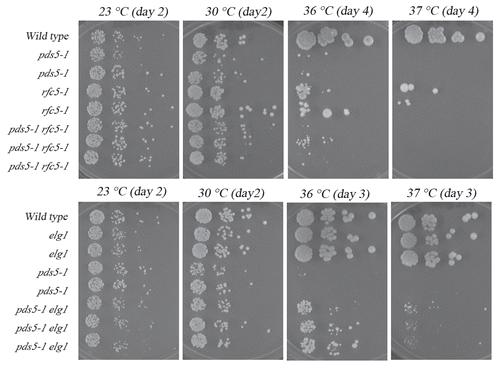
Figure 6 Possible roles for the small Rfc5p subunit in directing RFC complex function. (A) Contrasting roles of wild-type RFCs in regulating Ctf7p/Eco1p function. Ctf18p and Elg1p (large blue and salmon balls, respectively) both uniquely associate with Rfc2p-Rfc5p (small white balls, Rfc5p in purple) and also with PCNA (green cylinder) to regulate Ctf7p/Eco1p-dependent sister chromatid pairing. RFC arrangements as depicted previously.Citation44,Citation45 (B) rfc5-1p mutant protein (small tpurple pentagon) may be deficient in binding Elg1p but not Ctf18p, suggesting a role for Rfc5p in specifying large subunit recruitment or activation. (C) A highly speculative model in which Ctf18p can independently bind Dcc1p and Ctf8p (independent of RFC small subunits) to promote cohesion. Alternatively, Rfc5p may play a sensory role—possibly in detecting chromatin contexts (possibly required for cohesin deposition) normally involved in cohesion establishment (not shown).
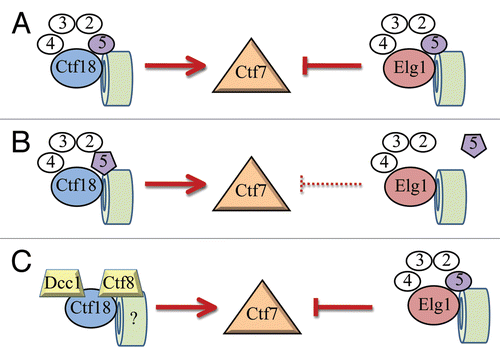
Table 1 Strains used in this study
Acknowledgements
The authors thank both Skibbens' and Cassimeris' lab members for comments throughout the course of this study. RVS is supported by an award from the National Institutes of General Medicine (1R15GM083269). Any opinions, findings and conclusions expressed in this study are those of the author(s) and do not necessarily reflect the views of the National Institutes of General Medicine.
References
- Indiani C, O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol 2006; 7:751 - 761
- Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci USA 2003; 100:10237 - 10242; PMID: 12930902
- Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol 2005; 25:5445 - 5455; PMID: 15964801
- Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol 2003; 13:1583 - 1595; PMID: 13678589
- Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J 2003; 22:4304 - 4313; PMID: 12912927
- Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci USA 2003; 100:9906 - 9911; PMID: 12909721
- Paulovich AG, Margulies RU, Garvik BM, Hartwell LH. RAD9, RAD17 and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 1997; 145:45 - 62; PMID: 9725831
- Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol 1998; 18:5485 - 5491; PMID: 9710632
- Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3 and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol 1999; 19:1136 - 1143; PMID: 9891048
- Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, et al. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J Biol Chem 2003; 278:50744 - 50753; PMID: 14530260
- Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 2004; 78:227 - 260; PMID: 15210332
- Skibbens RV. Mechanisms of sister chromatid pairing. Int Rev Cell Mol Biol 2008; 269:283 - 339; PMID: 18779060
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 2008; 24:105 - 129; PMID: 18616427
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science 2008; 321:566 - 569; PMID: 18653894
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 2008; 321:563 - 566; PMID: 18653893
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 2008; 31:143 - 151; PMID: 18614053
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev 1999; 13:307 - 319; PMID: 9990855
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 1999; 13:320 - 333; PMID: 9990856
- Skibbens RV. Buck the establishment: reinventing sister chromatid cohesion. Trends Cell Biol 2010; 20:507 - 513; PMID: 20620062
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC (Ctf18p, Ctf8p, Dcc1p): An alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 2001; 7:959 - 970; PMID: 11389843
- Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 2001; 21:3144 - 3158; PMID: 11287619
- Kenna MA, Skibbens RV. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different Replication Factor C Complexes. Mol Cell Biol 2003; 23:2999 - 3007; PMID: 12665596
- Maradeo ME, Skibbens RV. The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS One 2009; 4:4707; PMID: 19262753
- Parnas O, Zipin-Roitman A, Mazor Y, Liefshitz B, Ben-Aroya S, Kupiec M. The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS One 2009; 4:5497; PMID: 19430531
- Skibbens RV. Establishment of sister chromatid cohesion. Curr Biol 2009; 19:1126 - 1132; PMID: 20064425
- Yao NY, Johnson A, Bowman GD, Kuriyan J, O'Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J Biol Chem 2006; 281:17528 - 17539; PMID: 16608854
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, et al. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 2006; 23:787 - 799; PMID: 16962805
- Milutinovich M, Unal E, Ward C, Skibbens RV, Koshland D. A multi-step pathway for the establishment of sister chromatid cohesion. PLoS Genet 2007; 3:12; PMID: 17238288
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 2009; 33:763 - 774; PMID: 19328069
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature 2009; 462:231 - 234; PMID: 19907496
- O'Donnell M, Jeruzalmi D, Kuriyan J. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr Biol 2001; 11:935 - 946; PMID: 11719243
- Aroya SB, Kupiec M. The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Repair 2005; 4:409 - 417; PMID: 15725622
- Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol 2000; 20:5888 - 5896; PMID: 10913172
- Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol 1998; 18:5485 - 5491; PMID: 9710632
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 2001; 104:397 - 408; PMID: 11239397
- Xu H, Boone C, Brown GW. Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 2007; 176:1417 - 1429; PMID: 17483413
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol 2000; 151:613 - 626; PMID: 11062262
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5p cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol 2000; 10:1557 - 1564; PMID: 11137006
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumolyated to promote the dissolution of cohesion. J Cell Biol 2003; 163:729 - 741; PMID: 14623866
- Noble D, Kenna MA, Dix M, Skibbens RV, Unal E, Guacci V. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism. Cell Cycle 2006; 21:2528 - 2536; PMID: 17102636
- Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J 2001; 20:5779 - 5790; PMID: 11598020
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol 2009; 19:492 - 497; PMID: 19268589
- Rose MD, Winston F, Hieter P. Methods in yeast genetics 1990; Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press
- Venclovas C, Colvin ME, Thelen MP. Molecular modeling-based analysis of interactions in the RFCdependent clamp-loading process. Protein Sci 2002; 11:2403 - 2416; PMID: 12237462
- Kim J, MacNeill SA. Genome stability: a new member of the RFC family. Curr Biol 2003; 13:873 - 875; PMID: 14614842