Abstract
It is well established that there is a dynamic relationship between the expanding tumor and the host surrounding tissue. Cancer-associated fibroblasts (CAFs), the most common cellular population found in the tumor microenvironment, supporting tumor growth and dissemination. Here, we set out to determine the factors that may be involved in dramatic alteration of gene expression pattern in CAFs, focusing on microRNA and transcriptional regulators. We established matched pairs of human CAFs isolated from endometrial cancer and normal endometrial fibroblasts. MicroRNA and mRNA analyses identified differential expression of 11 microRNAs, with miR-31 being the most downregulated microRNA in CAFs (p=0.007). We examined several putative miR-31 target genes identified by microarray analysis and demonstrated that miR-31 directly targets the homeobox gene SATB2, which is responsible for chromatin remodeling and regulation of gene expression, and was significantly elevated in CAFs. The functional relevance of miR-31 and SATB2 were tested in in vitro models of endometrial cancer. Overexpression of miR-31 significantly impaired the ability of CAFs to stimulate tumor cell migration and invasion, without affecting tumor cell proliferation. Genetic manipulation of SATB2 levels in normal fibroblasts or CAFs showed that, reciprocally to miR-31, SATB2 increased tumor cell migration and invasion, while knock-down of endogenous SATB2 in CAFs reversed this phenotype. Introduction of SATB2 into normal fibroblasts stimulated expression of a number of genes involved in cell invasion, migration and scattering. These findings provide new insights into tumor-stroma interaction and document that miR
Introduction
The tumor microenvironment plays an important role in the development and progression of cancer.Citation1,Citation2 Epithelial carcinomas are surrounded by desmoplastic stroma consisting of fibroblasts, endothelial cells, pericytes, immune cells and various bone marrow-derived progenitor cells, including mesenchymal stem cells (reviewed in refs. Citation1 and Citation2). Although these stromal cells of the tumor microenvironment do not undergo malignant transformation, changes do arise in their gene expression profiles and, consequently, in their function that distinguish them from their normal counterparts.Citation3,Citation4
Fibroblasts are the most common cell type found in the tumor microenvironment.Citation5–Citation9 These cancer-associated fibroblasts (CAFs) are responsible for the synthesis of proteins involved in the remodeling of the extracellular matrix (ECM) as well as secretion of growth factors that regulate tumor cell proliferation, survival and dissemination.Citation7,Citation10 Using a mouse prostate tumor xenograft model, it has been shown that fibroblasts isolated from human prostate cancer, but not from normal prostate tissue, induce an oncogenic transformation of the non-tumorigenic immortalized prostate epithelial cells.Citation11,Citation12 Similarly, co-injection of fibroblasts isolated from invasive breast carcinomas with breast cancer cells enhanced tumor growth and produced highly vascularized tumors when compared to co-injection with normal breast fibroblasts isolated from the same patient or from other patients undergoing reduction mammoplasties.Citation13
CAFs mediate tumor promotion and dissemination through multiple mechanisms based on the secretion of growth factors (HGF, IGF, FGF, EGF, Wnt and TGFβ) and extracellular matrix-degrading metalloproteinases (MMPs) (reviewed in refs. Citation2 and Citation14). They also secrete stromal cell-derived factor 1 (SDF1), which stimulates tumor angiogenesis by recruiting endothelial progenitor cells and enhances tumor cell proliferation through its interaction with the CXCR4 receptor expressed by epithelial cancer cells.Citation13 In addition, MMPs, secreted by CAFs, are believed to play a major role in metastasis by activating the latent growth factors residing in the extracellular matrix (reviewed in ref. Citation7). MMP3, for instance, may directly cleave the extracellular domain of E-cadherin, a cell-cell adhesion molecule, inducing cancer cell metastasis.Citation15 Stroma-specific signaling through PTEN-Ets2 has been identified as a critical pathway involved in initiation and progression of mammary tumors.Citation16
Despite their ability to promote tumor growth, CAFs isolated from human carcinomas are not tumorigenic when injected into immunocompromised mice. Their growth characteristics are quite similar to normal fibroblasts and they begin to senesce after about 15 population doublings in vitro.Citation12,Citation13 Initially, several reports indicated a high incidence of chromosomal abnormalities in CAFs, including regions of loss of heterozygosity and mutations in tumor suppressor genes p53 and PTEN.Citation17–Citation20 However, recent analyses using array CGH or SNP arrays indicate that genetic alterations are extremely rare in CAFs.Citation21,Citation22 Therefore, dramatic alteration in gene expression may result from epigenetic mechanisms, either pre-existing in recruited bone marrow-derived cells, or initiated in resident fibroblasts by tumor cells, inflammation or tissue hypoxia.Citation23,Citation24
The tumor promoting functions of CAFs, in combination with their relative genetic stability, make them an attractive target for the development of novel therapeutic interventions. The experiments reported in this manuscript were designed to identify factors involved in the CAF phenotype, particularly those involved in gene transcription and translation. MicroRNAs are especially appealing because of their role as pleiotropic regulators of gene expression. Here, we report the differential expression of regulatory micro- and mRNAs in the CAFs derived from human endometrial cancer versus paired normal endometrial fibroblasts (NF) and demonstrtate that miR-31 downregulation in CAFs results in increased tumor cell motility, which is, in part, mediated by its direct targeting the homeobox gene SATB2.
Results
Primary fibroblast cultures.
We initially started the microRNA and mRNA analysis with ten fibroblast cell lines established from normal endometrium or endometrial cancer of five patients. Four of these cancer cases were classified as endometrioid with one case of clear cell histology. Three patients had grade III and two had grade II cancers. shows the clinicopathological data for the samples used.
The purity of the cultured fibroblast cells was verified by immunostaining using vimentin (mesenchymal cell marker) and cytokeratin (epithelial cell marker) (Sup. Fig. 1). All cell cultures were vimentin-positive and 91% to 100% cytokeratin negative. Several proteins known to be induced in CAFs (8) were tested by western blotting, and we observed increased amounts of Fibroblast-Specific Protein 1 (FSP1 or S100A4) and Fibroblast Activation Protein (FAP) (). In agreement with the previous reportCitation25 all established fibroblast cell lines expressed high levels of α-smooth muscle actin.
To exclude the possibility that CAFs may contain endometrial cancer cells that underwent an epithelial-to-mesenchymal transition and acquired mesenchymal cell markers, we performed karyotypic analysis on several CAF cell lines. While carcinoma cells usually have 20 to 50 different chromosomal aberrations, CAF cells showed a stable diploid karyotype. In addition, all fibroblast cell lines were injected subcutaneously into NOD. SCID/NCr mice at two different concentrations and observed for 120 days. We found no signs of neoplastic growth in these experiments.
To further characterize CAF's effect on tumor cells, we tested their ability to stimulate growth of endometrial cancer cells in co-culture experiments. Luciferase-expressing NCI-EC1 endometrial carcinoma cells (further called EC1) were plated together with equal numbers of CAFs or NFs. The cell mixture was propagated in medium with 2% serum for two days to allow fibroblasts to precondition the medium, and then serum was added to a final concentration of 5%. The results show that EC1 cells grew faster when co-cultured with CAFs than with corresponding NFs (). To analyze the effect of CAFs on tumor cells motility, we performed Transwell migration or matrigel invasion experiments. Fibroblasts, CAFs or NFs, were plated in low serum (2% FBS) medium, and Boyden chambers containing EC1 cells in serum-free medium were inserted the next day. In a parallel experiment, we determined that CAFs and NFs do not proliferate in the media with low serum and therefore there is no bias in the number of fibroblasts during the course of the experiment. and E demonstrate that both migration and invasion of EC1 cells were stimulated by CAFs as compared to normal fibroblasts.
Next, we tested the in vivo effect of CAFs on endometrial cancer cell growth. EC1 cells expressing the luciferase gene were injected alone or co-injected either with mouse embryo fibroblasts (MEF), human endometrial normal fibroblasts or endometrial CAFs. and G show images of mice and graphical presentation of these experiments at day 50 after subcutaneous injection. We observed a statistically significant increase in luciferase activity when EC1 cells were co-mingled with CAFs compared to other fibroblasts or tumor cells alone.
In summary, our primary cultures of CAF express fibroblastspecific markers, possess a normal karyotype, senesce after 12–15 passages, and CAFs stimulate tumor cell proliferation, migration and invasion when compared to their normal counterparts and promote tumor growth in vivo.
MicroRNA profiling.
The recent discovery of the regulatory role of microRNAs in different aspects of cancer progression prompted us to explore the microRNA signature of CAFs. Using a multispecies microarray chip, we identified 11 human microRNAs that were differentially expressed in CAFs compared to NFs (). Several microRNAs from other species showed statistically significant downregulation in endometrial CAFs marked by numbers in . As the degree of conservation of mature microRNA between species is very high, we determined the identity of those probes and found that three of them represented miR-31 homologs (). Probe 1093 recognizes C. elegans' miR-72, also a homolog of human miR-31.Citation26 Therefore, four different probes (including human miR-31) showed downregulation of miR-31 in CAFs. Among the seven upregulated microRNAs, three belong to the same cluster located on Xq26.3 (miR-503, miR-424 and miR-542-3p) and, therefore, are likely to be transcribed as a single transcriptional unit. To validate the differential expression of microRNAs in CAFs, we established ten more fibroblast cell lines derived from five additional patients. Overall, 20 cell lines, ten CAFs and ten respective NFs were subjected to stem-loop quantitative RT-PCR that detects expression of the mature microRNAs and demonstrated good correlation with the microarray data ().
mRNA microarray analysis.
To identify putative microRNA target genes, we carried out mRNA microarray analysis of RNA isolated from CAFs and NFs. MicroRNAs repress expression of their target genes by recognition of the specific sites in the 3′-untranslated region of mRNA and then by inhibiting their translation or inducing mRNA degradation. It has been demonstrated recently that mammalian microRNAs act predominantly by reducing mRNA stability,Citation27 that indicates high probability to reveal microRNA targeted molecules by transcriptional profiling.
The microarray data confirmed that only fibroblast-specific markers were expressed in these cells. The genetic markers indicative for epithelial cells, endothelial cells, macrophages or leukocytes were not detected (Sup. Table 1). Analysis of microarray data demonstrated enrichment in pathways that were described to be activated in CAFs. Quantitative real-time PCR confirmed an upregulation of a number of growth factors and MMPs that were previously detected in CAFs from different tumor types. Indeed, all endometrial CAFs showed increased levels of fibroblast-specific protein FSP1,Citation28 IGF2,Citation29 MMP3Citation15 and TGFβ2.Citation30 The list of all genes differentially expressed in endometrial CAFs selected with significant analysis of microarrays using FDR5, p < 0.01 and with a cutoff of more than two-fold, is provided in Supplemental Table 2.
We next asked if there was an enrichment in predicted target genes in our list of mRNAs differentially regulated in CAFs. We found that five out of 11 microRNAs showed statistically significant enrichment in their target genes (). This figure presents the number of predicted targets that negatively correlated with their corresponding microRNA and shows enrichment at p < 0.05 (Fischer's exact test).
Function of miR-31 in fibroblasts.
MiR-31 showed the highest degree of downregulation and was represented by multiple probes in our set. To test the functional relevance of miR-31 suppression, we first asked if loss of miR-31 in endometrial CAFs affects their ability to stimulate tumor cell growth and metastasis. We stably overexpressed the precursor of miR-31 or the control lentiviral vector containing GFP and the zeocin selection marker in CAFs from two different patients. After selection, more than 95% of cells were GFP-positive. The conditioned medium from fibroblasts overexpressing miR-31 reduced the migration and matrigel invasion of EC1 cells ( and B). However, the luciferase expressing endometrial cancer cell line EC1 showed no difference in growth rates when co-cultured with fibroblasts expressing miR-31 or vector control ().
SATB2 is a direct target of miR-31.
We previously found that miR-31 target genes have a high probability of being present in the list of differentially regulated genes generated from the mRNA expression analysis (). When we selected all miR-31 predicted targets (TargetScan, Release 4.0) from the mRNA microarray data, we found that all of them were upregulated in CAFs when miR-31 was downregulated (). Among the predicted targets, the SATB2 homeobox gene had the highest increase in its expression. SATB2 is a nuclear matrixattachment protein, involved in chromatin remodeling.Citation31,Citation32 In addition, through interaction with other transcription factors, it stimulates a cooperative increase of the transcription activation of their target genes. Quantitative RT-PCR of SATB2 and western blot analysis confirmed the higher expression in CAFs ( and C). The antibodies used for this analysis were specific for SATB2 and showed no crossreactivity with recombinant SATB1 protein (Sup. Fig. 2). We also observed a good negative correlation between SATB2 protein expression and miR-31 (compare and ).
The Target Scan database predicts two binding sites for miR-31 in the 3′UTR of SATB2 (). To address whether SATB2 is a direct target of miR-31, we cloned a 1.9 kB 3′UTR fragment containing both putative binding sites into the pMIRReport vector downstream of the luciferase gene. The reporter construct or empty vector control were co-transfected with either miR-31 mimic or a non-targeting control RNA into HeLa cells. We found that co-transfection with miR-31 did not affect the luciferase activity of the original empty vector control, while coexpression of miR-31 with the reporter containing SATB2 3′UTR reduced the luciferase activity (). To identify whether both predicted binding sites are functional, we cloned each of them separately into the same reporter vector. To prove the specificity of the miR-31 effect, we also deleted several base pairs in the seed regions of the miR-31 binding sites. The results show that each of the wild-type binding sites (but not the mutated sequence) is capable of reducing luciferase activity when co-transfected with miR-31 (). These data confirm that SATB2 is a direct target of miR-31.
We further checked whether overexpression of miR-31 would reduce endogenous SATB2 mRNA and protein levels. CAFs were transfected with miR-31 or negative control and SATB2 transcript levels were tested with quantitative RT-PCR (). MiR-148a is not predicted to target SATB2 and was used as a negative control. This experiment shows that overexpression of miR-31, but not a non-targeting negative control or miR-148a, resulted in reproducible and statistically significant reduction of SATB2 mRNA. To further confirm the repression of SATB2 protein, we overexpressed miR-31 in five selected CAF cell lines and found a profound effect of miR-31 on SATB2 protein levels ().
We also tested whether miR-31 targets ELAVL1 (also known as HuR), a gene known to interact with the 3′UTRs of mRNAs and involved in the control of mRNA stability or microRNA function.Citation33,Citation34 However, unlike SATB2, the 3′UTR fragment of the ELAVL1 gene cloned into the luciferase reporter vector was not affected by miR-31 co-expression (Sup. Fig. 3).
Function of SATB2 in cancer fibroblasts.
To test the role of SATB2 tumor cell migration or invasion, we performed a similar set of experiments using normal fibroblasts where the expression levels of SATB2 are lower than in CAFs. Overexpression of SATB2 stimulated migration and invasion (Matrigel) of endometrial cancer cells in Transwell migration assays ( and B). As with miR-31, we did not detect any change in growth rates of the EC1 endometrial cancer cells in the presence of fibroblasts overexpressing SATB2 protein (). We also used the reverse approach wherein we suppressed the endogenous SATB2 gene in CAFs by stable expression of a lentiviral construct delivering shRNA. shows that the migration of tumor cells towards SATB2-depleted CAFs was diminished compared to the same fibroblasts expressing a non-silencing control vector. The best shRNA construct we tested downregulated SATB2 mRNA level by 60% (), leaving a significant amount of gene expression intact, possibly explaining the modest effect of shSATB2 on endometrial cancer cell migration. shows SATB2 protein levels in cells expressing shSATB2 as compared with normal or cancer fibroblasts from the same patient. We also performed western blotting on normal fibroblasts overexpressing SATB2 protein to ensure that the levels of the ectopically expressed protein were close to physiological levels. For the comparison we included the data for patient 4, where the same amount of protein was run in the same gel and exposed for the same duration ().
To ascertain whether genes regulated by SATB2 in fibroblasts corroborate the increase in tumor cell motility, we studied differential gene expression in normal fibroblasts ectopically expressing SATB2 at levels similar to levels in CAFs. Three pairs of NF expressing either empty vector or SATB2 were subjected to microarray analysis. We first selected the differentially expressed genes that may provide paracrine signaling by fibroblasts, those that are secreted or localized on the plasma membrane. This set of genes was analyzed by IPA (Ingenuity Pathway Analysis) software tool. The results of this analysis showed that “Cellular movement” was the network with the highest score (). Analysis of all genes induced or suppressed by SATB2 more than two-fold also showed “Cellular Movement” as a top cellular function (). The subsections of this network include invasion of cells (p = 9.10E-07), migration of fibroblasts (p = 3.7E-03), scattering of cells (p = 1.95E-03), etc.
In summary, our data indicate that expression of miR-31 in fibroblasts suppresses tumor cell motility and invasion, at least in part, by targeting the SATB2 homeobox gene.
Discussion
The cellular components of the microenvironment appear to depend on alterations in transcriptional regulation, epigenetic modifications, chromatin structure and genome organization to manifest their highly specific phenotypes. Therefore, we hypothesized that microRNAs play an important role in regulation of specific genes present in the cells of the microenvironment that play a central role in the initiation and progression of human cancers. The experiments reported here demonstrate that nine out of ten pairs of human endometrial cancer fibroblasts showed diminished levels of the miR-31 microRNA. miR-31 has been identified previously as a differentially expressed marker in several types of human epithelial cancers. Several investigators have reported upregulation of miR-31 in colon cancerCitation35–Citation38 or in squamous cell carcinoma of the tongue,Citation39 as well as downregulation in breast cancer,Citation40 gastric cancerCitation41 and urothelial carcinomasCitation42 or aggressive forms of malignant mesothelioma.Citation43 Valastyan and co-authorsCitation44 demonstrated the loss of miR-31 in metastatic breast cancer cell lines and breast cancer patients. Using mouse xenograft models, the authors identified the role of miR-31 in suppression of breast cancer metastasis in mice without affecting tumor cell growth. These findings together with those presented here suggest that in order to metastasize, tumor cells suppress miR-31 expression in epithelial tumors and surrounding stromal cells.
Recently caveolin-1 deficiency has been implicated in the aggressiveness of breast cancer stromal fibroblasts,Citation45 and bone marrow-derived stromal cells from Cav-1(-/-) mice had elevated levels of miR-31.Citation46 However, it would be interesting to find out the effect of tumor cells on CAFs in Cav-1-negative background, since it is well known that that the interactions between CAFs and tumors are bidirectional and CAF signature depends not only on the tumor type but also on tumor progression.
Interestingly, miR-31 is localized at 9p21, in close proximity to the p16 CDK inhibitor, the well-known tumor suppressor locus CDKN2A that is frequently disrupted in a variety of human cancers. Since miR-31 is located less than 0.5 Mbases from p16, it would be expected to be lost together, and homozygous codeletions of miR-31 have been confirmed for several cases of urothelial carcinomas.Citation42 While the mechanism of miR-31 downregulation in CAFs has not been addressed in our studies, it is known that miR-31 is localized within the putative protein coding gene LOC554202.Citation47 Microarray data show that this gene was similarly downregulated more than four-fold in the endometrial cancer CAFs (p = 0.0139). Together these data imply that the two genes are likely to be co-transcribed from the same promoter.
Analysis of predicted miR-31 targets that matched the differentially expressed mRNAs demonstrated that all of the predicted target genes were upregulated in CAFs (). Several genes in the list are actually involved in actin filament organization and anchorage-independent growth, which may explain the reduced chemoattractant properties of cells overexpressing miR-31. Two other genes, SATB2 and ELAVL1, are involved in global chromatin remodeling and regulation or transcript stability, respectively. Both of these proteins may be responsible for the major changes observed in global gene expression. ELAVL1, also known as HuR, is an RNA binding protein that regulates transcript stability by binding to AT-rich regions in the 3′UTRs of its target mRNA. Overall changes in gene expression after modulation of the HuR gene in a breast cancer cell line showed that it promotes a more tumorigenic phenotype.Citation34 Furthermore, by interaction with its target genes' 3′UTR, HuR is able to interfere with microRNA function, likely by preventing the binding of microRNAs to their target genes.Citation33 Therefore, HuR gene upregulation in cancer-associated fibroblasts is a novel and interesting observation and worth pursuing in the future.
Of all putative miR-31 target genes, SATB2 showed the highest degree of upregulation in CAFs. The experiments reported here show that it is regulated by miR-31 through two binding sites in its 3′UTR. Both miR-31 and SATB2 affect migration and invasion of endometrial cancer cells but in opposite directions; miR-31 suppresses and SATB2 stimulates tumor cells migration/invasion. Prior to this study, SATB2 protein had not been implicated in tumorigenesis. However, its only family member, SATB1, is overexpressed in highly metastatic breast cancer cells, and its expression levels have high prognostic value for detection of aggressive forms of breast cancer.Citation48 SATB1 and SATB2 comprise a family of nuclear matrix-attachment proteins that recognize AT-rich DNA sequences at the base of looped-out chromatin, where attachment to the nuclear matrix occurs, and are involved in chromatin condensation, interaction with other chromatin remodeling complexes and regulation of transcription.Citation31,Citation49–Citation51
Haploinsufficiency in SATB2 in humans correlates with cleft palate.Citation32 In mice, targeted knockout of SATB2 results in multiple craniofacial abnormalities and perinatal lethality.Citation52,Citation53 A more detailed study found that SATB2 functions in skeletogenesis by driving osteoblast differentiation.Citation53 It does so by inhibiting the expression of several homeobox genes and activating Runx2 and ATF4 transcription factors. The SATB2-deficient osteoblasts show diminished levels of expression of Matrix Metalloproteinase 3 (MMP3) and increased expression of Tissue Inhibitor of Metalloproteinases 3 (TIMP3), implying direct or indirect regulation of these genes by SATB2. Similar to osteoblasts, endometrial CAFs express elevated levels of IGF2, Runx2, SATB2 and MMP3 as well as lower levels of TIMP3. Both increased MMP3 and IGF2 and suppression of TIMP3 were previously implicated in tumor-mediated tissue remodeling, leading to a more aggressive phenotype.
In conclusion, we characterized one particular pathway, miR-31-SATB2, that is partially responsible for CAF-mediated migration of tumor cells. This pathway does not affect tumor cell growth, which is also a well documented property of CAFs. We are currently trying to establish a mouse xenograft model to study metastatic spread of endometrial cancer cells that will allow us to confirm the results in vivo. More study is needed to evaluate the function of other microRNAs disregulated in CAFs. Understanding of the molecular mechanisms involved in CAF-mediated promotion of cancer growth and metastasis may represent promising targets for therapeutic intervention.
Materials and Methods
Cell lines.
Fresh tissues for the development of cell lines were obtained from patients undergoing hysterectomy for endometrial cancer. Tumor samples were annotated with clinical and pathological information but de-identified with respect to patient identifiers. The protocols for obtaining human tissues were approved by the Institutional Review Boards of the University of Virginia and the National Cancer Institute. Histologically normal appearing peritumoral tissue was separated from the tumor by the pathologist responsible for the case review. The pathologist removed normal tissue as distant as possible from the primary tumor and labeled such as normal tissue. Paired samples (normal and tumor) from each patient were transported to the NCI in DMEM medium containing antibiotics and antimycotics. Tissues were cut with a scalpel into pieces 1–2 mm3, placed on plastic dishes and covered with DMEM (high glucose) tissue culture media, supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and Penicillin/Streptomycin (Invitrogen). Cultures were incubated in humidified air containing 5% CO2 at 37°C. Fresh medium was added daily until tissue fragments were attached to the plastic.
Following tissue attachment (usually 2–3 days), the culture medium was changed twice a week for the next 2–3 weeks. Under these conditions, fibroblasts were explanted from tissue fragments while other cells were mostly retained within the tissue. Fibroblasts formed multiple dense colonies that spread out on the culture dish. After 2–3 weeks, cultured cells were briefly trypsinized and re-plated into T25 culture flasks as passage 1. After reaching confluence, the cultured cells were split 1:2 every 3–4 days. All fibroblasts used in experiments were between passage 4 and 9.
The NCI-EC1 (EC1) endometrial cancer cell line was established from a patient with stage IIIC, grade 3 endometrioid endometrial cancer (stage 3C). Cytogenetic analysis demonstrated that the majority of cells were tetraploid with multiple chromosomal rearrangements. EC1 cells were cultured in DMEM (high glucose) supplemented with 10% fetal bovine serum (Invitrogen) and Penicillin/Streptomycin (Invitrogen) under standard culture conditions.
All cells were routinely checked for mycoplasma contamination using MycoAlert Kit (Lonza).
Immunocytochemistry.
Fibroblasts at passage 3 were grown on cover slips, fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 for 4 min and immunostaining was performed with anti-Vimentin antibody (Dako, dilution 1:200) or anti-Cytokeratin antibody (Dako, dilution 1:100) using DakoCytomation EnVision + System according to the manufacturer's instructions.
Microarray analysis.
Total RNAs were purified from fibroblasts at passage 3 by Trizol Reagent (Invitrogen). 5 µg of total RNA were labeled and hybridized to an Affymetrix microarray chip HG-U133Plus2.0 according to manufacturer's instructions. The raw data were normalized using the GC-RMA algorithm, and differentially expressed genes were generated using the significance analysis of microarray (SAM) method. The cut-offs were set up at FDR = 5 and p-value <0.01 from a two-sided t-test. The SLEPR method, specifically developed to identify signaling pathways in gene expression analysis,Citation54 was used to analyze pathway enrichment.
MicroRNA analysis.
Total RNA was isolated from fibroblasts at passage 3, enriched with microRNA, labeled with Alexa Fluor-5 or Alexa Fluor-3 and hybridized to NCode Multi-Species miRNA microarray from Invitrogen, which targets all of the miRNA species in Sanger mirBase Release 9.0. The arrays were analyzed using the Sign Test statistical method. The average consistent fold change within the pattern was calculated using spots that show an increase (or decrease) in expression in CAFs versus NFs without inclusion of spots that did not follow the pattern. MicroRNAs that were increased or decreased by more than 1.5 fold at p-values less than 0.05 were considered significant and are presented in this publication. MicroRNAs from non-human species labeled by number-only probes can be found at orf.invitrogen.com/ncode.
Predicted target genes for microRNAs were obtained using the TargetScan release 4.0 database. To assess the predicted target enrichment for genes negatively correlated with an intended miRNA, correlation coefficients were calculated for each probe of a given miRNA, with each probe of mRNA on the miRNA chip and the mRNA chip, respectively. The enrichment levels were assessed using Fisher's exact test based on a 2 × 2 contingency table (whether a gene is a predicted target or not versus whether a specific gene had a good negative correlation with the intended miRNA at correlation p-value < 0.05).
Plasmids.
To test direct targeting of SATB2 by miR-31, we cloned its 3′UTR into pMIR-Report vector (Ambion) by RT-PCR using primers: forward primer 5′-CTT GCT ACG TTC CGT TAT TCA ATT TGT CAT TAC TG-3′ and reverse primer 5′-CAT ATT TAT TGA ATG CGT TTA TTT TAA CAA CC-3′. Potential miR-31 binding sites in the 3′UTR of SATB2 were identified using the TargetScan database (MIT, www.targetscan.org). Each of the two predicted binding sites were cloned separately in pMir-Report vector into Spe1-HindIII restriction sites using synthetic oligonucleotides: site 1 (S1) (sense strand) 5′-GTA GTA GGT ATT TTT CAA TGC TAA GTC TTG CCT TTT ATT TTT TAA TTT CAC TGC CAA-3′ and site 2 (S2) (sense strand): 5′-CTA GTG TGG GCA TTT TAG CCT GTG GTC TTG CCA GAT CTT TGC GAA TTA CAA TGC ATA-3′. To produce mutated miR-31 binding sites the seed regions (underlined in the oligos above) were deleted using QuickChange Site-directed Mutagenesis Kit (Stratagene). All plasmids were verified by sequencing.
Luciferase reporter assay.
105 HeLa cells were first transfected with microRNA or non-silencing controls (Dharmacon) using HiPerfect Transfection Reagent (Qiagen) followed by co-transfection with luciferase reporter constructs and pCMV-lacZ using Effectene Transfection Reagent (Qiagen) on the next day. Briefly, 750 ng of microRNA or non-silencing control (final concentration 50 nM) were mixed with 6 µl of Transfection Reagent in a total volume of 100 µl. The complex was formed for 10 min at room temperature and mixed with 105 HeLa cells in a total volume of 1.2 ml of complete medium. The mixture was plated in 12-well plates in triplicate for each condition. Cells were transfected the next day with 0.2 µg of firefly luciferase reporter constructs and 0.1 µg of CMV-lacZ plasmid using Effectene Transfection Reagent according to manufacturer's instructions. After 24 hours, cells were lysed in 250 µl of Passive Lysis Buffer (Promega) and 20 µl were used to measure luciferase activity with the Luciferase Assay System (Promega), or 50 µl were used to measure β-galactosidase activity using the β-Gal Assay Kit (Invitrogen).
Quantitative RT-PCR.
Total RNA was purified using Trizol Reagent (Life Technologies), and 1 µg of total RNA was reverse transcribed in 50 µl reaction using TaqMan Reverse Transcription Reagents (Applied Biosystems Inc.). 5 µl of the reverse transcribed cDNA were subjected to PCR according to Applied Biosystems technical recommendations. TaqMan probes were purchased from Assay-on-Demand (Applied Biosystems). Three replicate reactions were run for each RNA sample.
Western blotting.
Nuclear proteins were purified using NE-Per Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) according to the manufacturer's instructions. 20 µg of nuclear protein were fractionated in a 10% SDS-PAGE gel and transferred to PVDF membrane (Immobilon-P, Millipore). The membrane was probed with anti-SATB2 antibody (Novus Biologicals) followed by washing and incubation with HRP-conjugated secondary antibody (Pierce) and developed using ECLTM reagents (Amersham Biosciences).
Cell transfection.
Fibroblasts were transfected with Non-Silencing control or miR-31 mimic (both from Dharmacon) using Hi-Perfect Transfection Reagent (Qiagen). Briefly, cells were trypsinized and counted, then 2 × 105 cells were plated in 1.9 ml of complete medium into a 60 mm plate. The transfection complex was prepared at the same time and consisted of 100 µl of serum-free medium, 6 µl of microRNA (20 µM) or nontargeting control (Dharmacon) and 18 µl of HiPerfect reagent. The mixture was incubated for 15 min at room temperature and added to the cells. The final concentration of microRNA was 60 nM. Cells with transfection complexes were incubated for 24–48 hours without medium change.
Stable expression of miR-31 (SBI), SATB2 (Gene Copoeia) or shSATB2 (Open Biosystems) was achieved by lentiviral transduction and selection with appropriate antibiotic.
Co-culture of fibroblasts with endometrial cancer cells.
EC1 endometrial cancer cells were transfected with a retroviral vector expressing the firefly luciferase gene (a kind gift from Dr. Girma Woldemichael, NCI at Frederick), and transfected cells were selected with G-418. For co-culture experiments, 104 cells per well were seeded in triplicate in 24-well plates with 2 × 104 fibroblasts (CAF or NF) in DMEM, supplemented with 2% FBS. At 4 hours, when cells attached to the plastic, the “0” time points were collected. Cells were cultured for an additional 48 hours in the medium with 2% FBS. Limited EC1 proliferation was observed during this 48 hr period. After 48 hours, an equal volume of medium supplemented with 10% FBS was added, and luciferase activity was determined every day for five consecutive days. Luciferase activity was measured using the Luciferase Assay System (Promega) after cells were washed with ice-cold PBS and lysed in 50 µl Cell Culture Lysis Reagent (Promega) for 5 min. 10 µl of the lysate were used for each of the three measurements. Each measurement was done in triplicates.
Transwell migration/matrigel invasion.
Fibroblasts were seeded at a density of 5 × 104 per well in 24-well plates. At 4 hours, when cells were attached to the plastic, the medium was changed to 2% FBS. The next day, NCI-EC1 endometrial cancer cells were trypsinized, counted and allowed to recover in complete medium for 2 hours at 37°C. Cells were washed with PBS, resuspended in serum-free medium, counted and 5 × 104 cells were placed in the Boyden chamber with the untreated membrane for migration assay or Matrigel-covered membrane for invasion assay. The migration and invasion assays were carried out for 24 and 48 hours, respectively. Cells were fixed and stained with Diff-Quik Stain Set (Dade Behring). The cells on the lower side of the membrane were counted by a different person in a blind fashion. All assays were performed in triplicate and experiments were repeated at least three times.
Mouse xenograft models.
EC1 endometrial cancer cells expressing luciferase (5 × 105) were co-mingled with 1.5 × 106 fibroblasts, mixed with Growth Factor Reduced Matrigel (BD Lifesciences) and injected subcutaneously into the right flank of 8-week-old female nude mice (BalbC/nude, Charles River). Images were taken weekly after injection of 3 mg of Luciferin (Caliper).
Figures and Tables
Figure 1 Characterization of fibroblast cell lines. (A) Clinicopathologic data of the samples used to produce fibroblasts. (B) Western blot analysis of fibroblasts (CAFs and NFs) with anti-Fibroblast Specific Protein 1 (FSP1) and anti-Fibroblast Activation Protein (FAP). N, NF; C, CAF. (C) CAFs stimulate growth of endometrial cancer cell line (EC1) compared to normal fibroblasts in co-culture experiments. (D and E) Conditioned media from CAFs stimulate EC1 cells matrigel invasion (D) and migration (E). Values in (C–E) represent average numbers for five pairs of fibroblasts ± SEM. (F) Images of mice 50 days after injection with EC1-luc cells co-mingled with either NFs or CAFs. (G) Quantification of tumor burden in mice injected with EC1-luc alone or in combination with either mouse embryo fibroblasts (MEFs), endometrial NFs or endometrial CAFs.
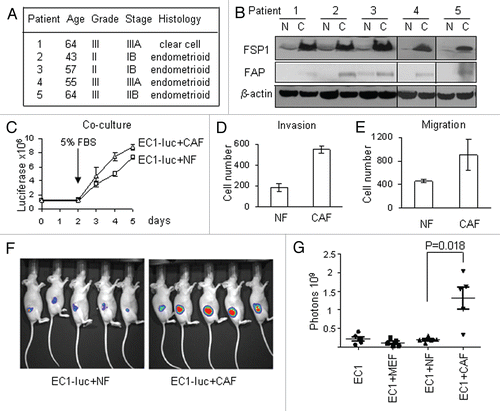
Figure 2 MicroRNA analysis of CAFs. (A) List of microRNAs differentially expressed in CAFs relative to normal fibroblasts. *The same cluster of microRNAs. #The same family of microRNAs. (B) The microRNAs recognized by the probes representing non-human species on the microarrays. (C) Stem-loop RT-PCR validations of microRNAs differentially expressed in CAFs. Black bars, NF; white bars, CAFs. (D) MicroRNA's predicted target enrichment.
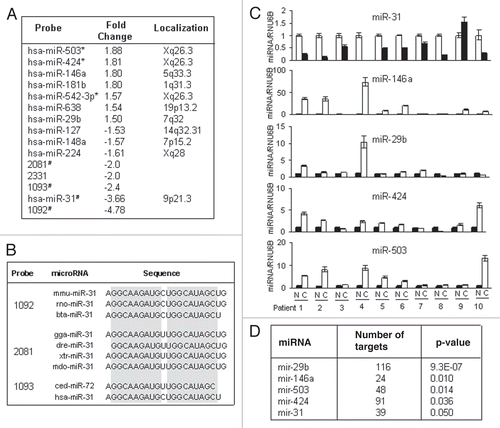
Figure 3 MiR-31 expression in fibroblasts decreases tumor cell migration and invasion. (A) Photographs depict the EC1 cancer cells migration towards the media preconditioned by CAFs stably transduced with miR-31 or empty vector (EV) lentiviral constructs in Transwell migration assays. (B) Quantification of EC1 cell migration or matrigel invasion is presented as mean ± SEM. Experiments were performed in triplicate and repeated at least three times using CAFs from two different patients. Representative experiment is shown. p values were obtained by paired t-test (*p < 0.05, **p < 0.01). (C) miR-31-transduced CAFs do not affect EC1 cell growth in co-culture experiments.
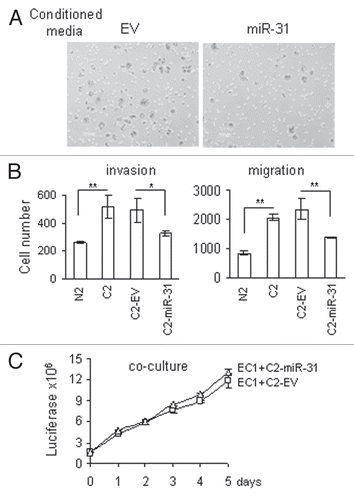
Figure 4 MiR-31-predicted target genes showed differential expression in CAFs. (A) Genes that matched TargetScan predicted genes in the list of genes generated by Significance Analysis of Microarray (SAM) method. Fold induction shows the ratio of expression in CAFs vs. normal fibroblasts. (B) Quantitative RT-PCR validations of SATB2 gene expression in ten pairs of fibroblasts. Black bars, NF; white bars, CAFs. Expression values for CAFs are presented relative to their respective NFs. (C) Western blot analysis of SATB2 protein expression.
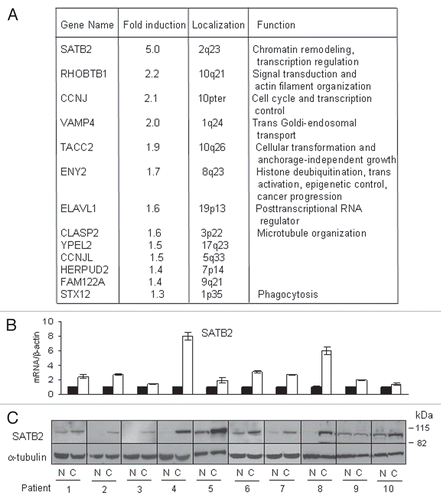
Figure 5 SATB2 is a direct target of miR-31. (A) Schematic representation of SATB2 mRNA and the positions of two predicted miR-31 binding sites (S1 and S2) in the 3′UTR. (B) Luciferase activity in HeLa cells after transfection with 1.9 kb SATB2 3′UTR reporter construct containing two binding sites and miR-31 (left part). Ctr, non-targeting control; EV, empty pMIR-Report vector. MiR-31 targets each of the binding sites, S1 (middle part) and S2 (right part) when cloned separately in the pMIR-Report vector. The effect of miR-31 was eliminated by mutations in its binding sites. The experiments were repeated at least three times with similar results, and the representative experiment is shown. (C) Quantitative RT-PCR of SATB2 transcript after overexpression of miR-31 or non-targeting control in CAFs. MiR-148a has been used as additional negative control. The results are mean values of relative mRNA levels normalized to beta-actin ± SEM. From at least three experiments. p-values were obtained by paired t-tests (*p < 0.01). (D) Western blot analysis of SATB2 protein downregulation in five pairs of CAFs after overexpression of miR-31. Endometrial CAFs were transfected with non-targeting control or miR-31 mimic, and three days later cells were collected for western blotting.
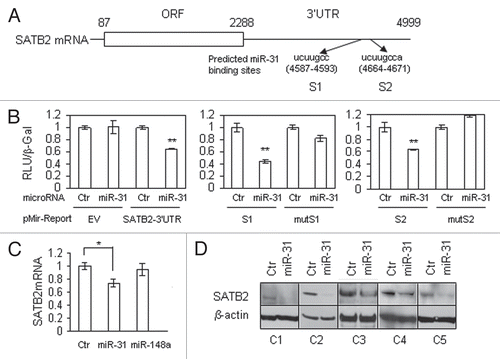
Figure 6 SATB2 stimulates endometrial cancer cell migration and invasion. Transwell migration and matrigel invasion experiments were performed using normal fibroblasts (N) transduced with SATB2 lentiviral construct. (A) Microphotographs show endometrial cancer EC1 cells migration towards media pre-conditioned with normal fibroblasts expressing empty vector (EV) control or SATB2. (B) Quantification of EC1 cells migration and invasion experiments were performed as in . (C) Co-culture of luciferase-labeled EC1 cells with normal fibroblasts expressing SATB2 or empty vector. (D) Knockdown of SATB2 in CAFs decrease their ability to stimulate EC1 cell migration. NS, non-silencing control. *p = 0.015. (E) Quantitative RT-PCR of SATB2 knockdown by stable expression of lentiviral vector with shSATB2. (F) Western blot analysis of SATB2 protein in CAFs overexpressing shSATB2 or normal fibroblasts with ectopic expression of SATB2 protein. N2 and C2, normal fibroblasts and CAFs, respectively, used for SATB2 knock-down. The last part shows endogenous levels of SATB2 in patient 4, which is comparable with the levels of SATB2 protein overexpressed in normal fibroblasts (patient 2). Same amounts of nuclear lysate were run in the same gel and exposed for the same time.
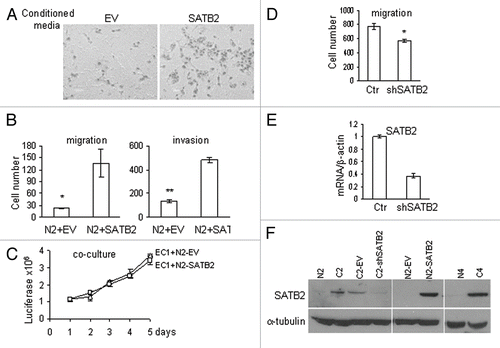
Figure 7 Expression of SATB2 in normal endometrial fibroblasts induces genes involved in cellular motility. (A) A majority of genes involved in paracrine signaling by fibroblasts (localized in extracellular space or plasma membrane) upregulated by SATB2 and the top network, designated as “cellular movement,” is presented. (B) Top five molecular and cellular functions enriched in SATB2 expressed normal fibroblasts. All genes with more than two-fold up or downregulation by SATB2 were used to assess the SATB2 functions.
Additional material
Download Zip (322.6 KB)Acknowledgements
We thank Dr. Girma Woldemichael for the kind gift of cells producing retrovirus with luciferase gene and Dr. Bradley Love (Invitrogen) for the help with microRNA array analysis. We are also grateful to Mrs. Hui Han (National Cancer Institute) for help with the cloning constructs and Ms. Susan Dalton (University of Virginia) for help with the sample collection. We thank Dr. Melinda Hollingshead, Ms. Jalpa Shah and Ms. Angelena Millione for help with fibroblast cultures in NOD. SCID mice. We thank Mr. Brian P. Hibler for critical reading of the manuscript. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The National Cancer Institute is accredited by AAALACi and follows the Public Health Service Policy on the Care and Use of Laboratory Animals. All animals used in this research project were cared for and used humanely according to the following policies: The US Public Health Service Policy on Humane Care and Use of Animals (1996); the Guide for the Care and Use of Laboratory Animals (NIH publication No. 86-23, 1985); and the US Government Principles for Utilization and Care of Vertebrate Animals Used in Testing, Research and Training (1985).
References
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer 2009; 9:239 - 252
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer 2006; 6:392 - 401
- Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet 2005; 37:899 - 905
- Polyak K. Breast cancer: origins and evolution. J Clin Invest 2007; 117:3155 - 3163
- Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420:860 - 867
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57 - 70
- Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol 2006; 1:119 - 150
- Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: Microenvironmental influences in the normal and malignant breast. Differentiation 2002; 70:537 - 546
- Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle 2006; 5:1597 - 1601
- Lorusso G, Ruegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem Cell Biol 2008; 130:1091 - 1103
- Hayward SW, Wang Y, Cao M, Hom YK, Zhang B, Grossfeld GD, et al. Malignant transformation in a nontumorigenic human prostatic epithelial cell line. Cancer Res 2001; 61:8135 - 8142
- Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res 1999; 59:5002 - 5011
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121:335 - 348
- Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature 2004; 432:332 - 337
- Lochter A, Galosy S, Muschler J, Freedman N, Werb Z, Bissell MJ. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol 1997; 139:1861 - 1872
- Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature 2009; 461:1084 - 1091
- Fukino K, Shen L, Matsumoto S, Morrison CD, Mutter GL, Eng C. Combined total genome loss of heterozygosity scan of breast cancer stroma and epithelium reveals multiplicity of stromal targets. Cancer Res 2004; 64:7231 - 7236
- Hill R, Song Y, Cardiff RD, Van Dyke T. Selective evolution of stromal mesenchyme with p53 loss in response to epithelial tumorigenesis. Cell 2005; 123:1001 - 1011
- Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet 2002; 32:355 - 357
- Moinfar F, Man YG, Arnould L, Bratthauer GL, Ratschek M, Tavassoli FA. Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res 2000; 60:2562 - 2566
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 2004; 6:17 - 32
- Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet 2008; 40:650 - 655
- Blagosklonny MV. Molecular theory of cancer. Cancer Biol Ther 2005; 4:621 - 627
- Polyak K, Kalluri R. The role of the microenvironment in mammary gland development and cancer. Cold Spring Harb Perspect Biol 2010; 2:a003244
- Czernobilsky B, Gabbiani G, Prus D, Lifschitz-Mercer B. Alpha-smooth muscle actin-positive myofibroblasts in endometrial stroma are not a reliable criterion for the diagnosis of well differentiated endometrioid adenocarcinoma in small tissue samples. Int J Gynecol Pathol 2001; 20:232 - 238
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, et al. The microRNAs of Caenorhabditis elegans. Genes Dev 2003; 17:991 - 1008
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466:835 - 840
- Cabezon T, Celis JE, Skibshoj I, Klingelhofer J, Grigorian M, Gromov P, et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int J Cancer 2007; 121:1433 - 1444
- Singer C, Rasmussen A, Smith HS, Lippman ME, Lynch HT, Cullen KJ. Malignant breast epithelium selects for insulin-like growth factor II expression in breast stroma: evidence for paracrine function. Cancer Res 1995; 55:2448 - 2454
- Nakagawa H, Liyanarachchi S, Davuluri RV, Auer H, Martin EW Jr, de la Chapelle A, et al. Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene 2004; 23:7366 - 7377
- Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev 2003; 17:3048 - 3061
- FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet 2003; 12:2491 - 2501
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006; 125:1111 - 1124
- Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, et al. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene 2008; 27:6151 - 6163
- Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 2006; 5:29
- Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 2009; 34:1069 - 1075
- Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2007; 72:397 - 402
- Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers 2009; 26:27 - 34
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 2008; 14:2588 - 2592
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008; 14:2348 - 2360
- Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 2009; 24:652 - 657
- Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, et al. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis and frequent homozygous losses of miR-31. Int J Cancer 2009; 124:2236 - 2242
- Ivanov SV, Goparaju CM, Lopez P, Zavadil J, Toren-Haritan G, Rosenwald S, et al. Pro-tumorigenic effects of miR-31 loss in mesothelioma. J Biol Chem 2010; 285:22809 - 22817
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009; 137:1032 - 1046
- Mercier I, Casimiro MC, Wang C, Rosenberg AL, Quong J, Minkeu A, et al. Human breast cancer-associated fibroblasts (CAFs) show caveolin-1 downregulation and RB tumor suppressor functional inactivation: Implications for the response to hormonal therapy. Cancer Biol Ther 2008; 7:1212 - 1225
- Pavlides S, Tsirigos A, Migneco G, Whitaker-Menezes D, Chiavarina B, Flomenberg N, et al. The autophagic tumor stroma model of cancer. Cell Cycle 2010; 9:3485 - 3505
- Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS ONE 2009; 4:e5279
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 2008; 452:187 - 193
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci 2005; 21:658 - 668
- Gyorgy AB, Szemes M, de Juan Romero C, Tarabykin V, Agoston DV. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur J Neurosci 2008; 27:865 - 873
- Szemes M, Gyorgy A, Paweletz C, Dobi A, Agoston DV. Isolation and characterization of SATB2, a novel AT-rich DNA binding protein expressed in development- and cell-specific manner in the rat brain. Neurochem Res 2006; 31:237 - 246
- Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, Tarabykin V. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am J Hum Genet 2006; 79:668 - 678
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, et al. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 2006; 125:971 - 986
- Yi M, Stephens RM. SLEPR: A sample-level enrichment-based pathway ranking method—seeking biological themes through pathway-level consistency. PLoS ONE 2008; 3:e3288