Abstract
mRNA polyadenylation and deadenylation are important processes that allow rapid regulation of gene expression in response to different cellular conditions. Almost all eukaryotic mRNA precursors undergo a co-transcriptional cleavage followed by polyadenylation at the 3' end. After the signals are selected, polyadenylation occurs to full extent, suggesting that this first round of polyadenylation is a default modification for most mRNAs. However, the length of these poly(A) tails changes by the activation of deadenylation, which might regulate gene expression by affecting mRNA stability, mRNA transport, or translation initiation. The mechanisms behind deadenylation activation are highly regulated and associated with cellular conditions such as development, mRNA surveillance, DNA damage response, cell differentiation and cancer. After deadenylation, depending on the cellular response, some mRNAs might undergo an extension of the poly(A) tail or degradation. The polyadenylation/deadenylation machinery itself, miRNAs, or RNA binding factors are involved in the regulation of polyadenylation/deadenylation. Here, we review the mechanistic connections between polyadenylation and deadenylation and how the two processes are regulated in different cellular conditions. It is our conviction that further studies of the interplay between polyadenylation and deadenylation will provide critical information required for a mechanistic understanding of several diseases, including cancer development.
Introduction
The production of mature mRNA requires the synthesis of a pre-mRNA by RNA polymerase II (RNAP II) and co-transcriptional processing of the nascent precursor by 5′ end capping, splicing and 3′ end processing.Citation1 The 3′ end of the mRNA is processed by the cleavage of the pre-mRNA at the polyadenylation site followed by the addition of a non-templated polyadenylated tail, which in mammalian cells is approximately 200–300 adenosine residues long.Citation2 Almost all eukaryotic pre-mRNAs undergo cleavage/polyadenylation at their 3′ ends, with the exception of histone mRNAs, which are cleaved but not polyadenylated.Citation3 The assembly of the cleavage/polyadenylation machinery requires specific signal sequences in the pre-mRNA as well as the assembly of a large number of protein factors. In normal mammalian cells, the cleavage step requires cleavage-polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factors I and II (CF I and II), RNAP II and poly(A) polymerase (PAP), while the subsequent poly(A)-tail synthesis depends on CPSF, PAP, symplekin and poly(A)-binding protein nuclear I (PABNI).Citation4,Citation5 The binding of PABN1 to the poly(A) not only stimulates PAP processivity but also prevents degradation of mRNAs and thereby preserves the length of the poly(A).Citation6 Although the assembly of all these factors is believed to be sufficient for 3′ processing to occur, additional protein factors have been identified as part of the mammalian 3′ end processing machinery.Citation7 For example, interconnections between the polyadenylation and the splicing/transcription machineries have been shown to play a role in the regulation of the specificity and efficiency of the cleavage/polyadenylation step.Citation8,Citation9 These additional factors have a role in the selection of alternative polyadenylation (APA) signals in most mRNAs, and in the activation of those signals in all the mRNAs. Cis-acting elements within the 3′ untranslated region (3′UTR), such as APA signals, microRNA (miRNA) target sites and AU-rich elements (ARE), play also an important role regulating 3′ processing. The relevance of these regulatory processes is highlighted by changes in the length of the 3′UTR of different mRNAs in cancer cellsCitation10,Citation11 and during cell differentiation.Citation12–Citation14 For better understanding of basic aspects of 3′ end formation and its regulation, we suggest the reading of a recent review by Mandel et al.Citation5
As no mechanism has been described yet that regulates the length of the poly(A) tail during the nuclear cleavage/polyadenylation reaction, it is easy to view this modification as a default mRNA processing step. However, once the tail has been synthesized the length of the poly(A) tail is subsequently modulated in what appears to be a highly regulated event, which outcome varies dependent on different cellular conditions. Thus, mechanisms involving poly(A) tail length control and degradation represent an additional checkpoint for regulation of eukaryotic gene expression.Citation4,Citation5,Citation15,Citation16 In this regard PAPs and deadenylases are the key catalytic entities that are required for proper regulation of poly(A) tail length. Evidence implies that deadenylases are pivotal in this process and hence deadenylase activities have gained considerable interest and a number of deadenylases have been identified and studied in detail.Citation17,Citation18 The best characterized deadenylases so far are the CCR4-POP2-NOT complex, which is a predominant deadenylase in all eukaryotes, the poly(A)-specific ribonuclease (PARN), which is a major deadenylase activity in mammals and poly(A) nuclease (PAN), which is involved in early steps of poly(A) tail metabolism. Studies have shown that deadenylases localize both in the nucleus and the cytoplasm and are involved in different cellular processes such as DNA damage response (DDR), non-sense mediated decay (NMD), cell cycle regulation, cell growth control, cell proliferation, inflammatory response, cell differentiation, etc.Citation19–Citation24 The role of deadenylation in these cellular processes is either to decrease the total mRNA levels, to regulate the levels of specific mRNAs through sequence elements present in their 3′UTR or to participate in modulating the length of the poly(A) tail. In addition to the deadenylases the multicomponent exosome participates in the turnover of both total mRNA levels and specific mRNAs.Citation25–Citation27 After deadenylation, depending on the cellular response, some mRNAs might undergo a new round of poly(A) tail extension and translation, while others might be degraded by the exosome.
How the poly(A) tail length of a particular mRNA and, consequently, its level of translation is determined has been a matter of investigation for many years. In this review we discuss mechanistic connections between polyadenylation and deadenylation in the nucleus () and cytoplasm () under different cellular conditions, as it has become clear that poly(A) tail length results from a balance between concomitant deadenylation and polyadenylation. Besides, we also focus our review on cis-acting sequence elements present in the 3′UTR that have been shown to participate in the regulation of polyadenylation/deadenylation of specific mRNAs ( and ). Such cis-acting sequences are recognized by miRNAs, ARE-binding proteins, polyadenylation factors or other RNA binding factors. In summary, this review provides a broader understanding of the interplay between polyadenylation and deadenylation in gene regulation, and discusses the implications of deregulation of these important steps of mRNA maturation in disease.
Nuclear Polyadenylation/Deadenylation
The nuclear polyadenylation and deadenylation machineries have been mechanistically implicated in mRNA surveillance and DDR. Nuclear degradation of mRNAs with mutations in their coding sequence as well as pre-mRNAs with defective splicing or 3′ end processing requires the exosome.Citation28–Citation40 The nuclear exosome also participates in the degradation of non-coding RNAs, including tRNA, rRNA, snRNA, snoRNA and cryptic non-protein coding transcripts generated by RNAP II.Citation41 In yeast, a nuclear mechanism of RNA surveillance has been discovered that involves not only deadenylation but also active polyadenylation by the TRAMP complex ().Citation26,Citation42,Citation43 The TRAMP complex is formed by two noncanonical PAPs, referred as Trf4p and Trf5p, a 3′ to 5′ RNA helicase (Mtr4p) and an RNA-binding protein (either Air1p or Air2p). Polyadenylation by the TRAMP complex adds a tail that is around 40 nucleotides long to the 3′ end of the targeted RNA and marks it for degradation. It has been shown that the CCR4-NOT complex, which includes the major yeast deadenylase CCR4, physically and functionally interacts with TRAMP and the nuclear exosome.Citation44 Interestingly, the TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6.Citation45 Rrp6 has exonucleolytic activity on aberrantly 3′ end processed transcripts and retains transcripts at sites of transcription where they are presumed to be degraded.Citation32,Citation39,Citation46 Supporting these observations, Milligan et al.Citation39 observed not only reduction in the levels of different mRNA species but also of truncated RNAs in yeast strains with a defective PAP. Further studies are required to determine if a homologue of the TRAMP complex, which includes polyadenylation/deadenylation mechanisms, exists in mammalian cells. Interestingly, a similar quality-control pathway for polyadenylated pre-mRNAs to those found in yeast has been described in the nucleus of mammalian cells.Citation47 This rapid mammalian deadenylation-dependent decay pathway for intronless polyadenylated transcripts involves the viral RNA element ENE, which is essential for the nuclear accumulation of a viral polyadenylated nuclear RNA. The ENE inhibits deadenylation and decay of RNAs, likely through intramolecular hybridization with the poly(A) tail.Citation48
Another example of mechanistic connections between polyadenylation and deadenylation machineries has been described in the DDR in mammalian cells ().Citation49 In this case, it is possible that the nuclear polyadenylation/deadenylation machineries might signal the degradation of prematurely terminated and erroneously processed transcripts, which could be generated by the arrest and degradation of RNAP II at sites of DNA damage. This process might represent a mechanism of nuclear mRNA decay that avoids the formation of potentially deleterious hazardous proteins. It has been shown that RNA 3′ end processing is strongly but transiently inhibited upon DNA damaging conditions.Citation50 Initially, it was described that 3′ end processing is inhibited after UV-induced DNA damage as a result of both the formation of the BARD1/CstF complexCitation50 and the proteasomal-mediated degradation of RNAP II.Citation51 However, the UV-induced inhibition of 3′ processing turned out to be mechanistically more complex than initially thought. For example, CstF1 can also interact with PARN upon UV treatment.Citation49 Based on the nature of the factors, it is possible to hypothesize that the PARN/CstF1 interaction might regulate mRNA turnover in different cellular responses. Consistent with this, the CstF1/PARN complex plays a role in inhibition of mRNA 3′ cleavage and activation of deadenylation upon DNA damage. Importantly, the tumor suppressor BARD1 also plays a role in the activation of deadenylation by PARN,Citation49 suggesting that the activation of deadenylation and the inhibition of 3′ mRNA cleavage are part of the same cellular response to DNA damage. Interestingly, the cap binding protein 80 (CBP80) also binds PARN inhibiting its deadenylase activity in untreated cells,Citation52 representing a possible mechanism by which PARN is recruited to the nascent pre-mRNA and ensuring that PARN does not degrade the poly(A) tail of pre-mRNAs. However, following UV-treatment, the CstF1/BARD1 complex can revert the CBP80-mediated inhibition of PARN activity, suggesting a functional link between the 5′ cap and the polyadenylation/deadenylation machinery in the DDR. Extending those studies, it was also shown that PARN along with CstF1/BARD1 complex participate in the regulation of endogenous transcripts during the DDR either by inhibition of 3′ cleavage or by activation of deadenylation.Citation49 Interestingly, nuclear PARN also plays a role decreasing the stability of ARE-containing mRNAs, such as c-myc, c-fos and c-jun, and keeping their levels low in non-stress conditions.Citation49,Citation53,Citation54 Then these short lived mRNAs increase transiently upon DNA damaging conditions and are involved in control of cell growth and differentiation.Citation55–Citation57
Supporting the role of PARN deadenylase in DDR, other deadenylases have also been functionally connected with factors involved in the DDR in yeast. For example, both PAN and the CCR4-CAF1 complex can interact with DNA damage response protein kinase DUN1 (Dun1), which is involved in the DNA damage-dependent induction of repair genes and in cell cycle arrest.Citation58,Citation59 The mRNA levels of the DNA repair gene RAD5 are regulated by a mechanism involving poly(A) tail length control. Dun1 and PAN complex act together in this deadenylation process, representing additional checkpoint targets in the regulation of DNA repair. In addition, the CCR4-NOT complex genetically interacts with different checkpoint pathwaysCitation59 and mutants in the deadenylase CCR4 have been found to be sensitive to DNA damage treatment.Citation60 Interestingly, CCR4-NOT and PAF1, which have mRNA 3′ end processing functions, are involved in the transcription-coupled repair pathway.Citation61 It is also possible that the exosome-associated deadenylase activity has a role in the degradation of incorrectly processed and defective transcripts generated during DDR. Rrp6, which is an mRNA surveillance factor, and its binding partner Lrp1, which is a DNA repair protein, participate not only in general mRNA degradation but also in specific mRNA degradation upon UV treatment.Citation46 Lrp1 is involved in DDR and requires Rrp6 for proper localization on target mRNAs under both DNA damaging and non-damaging conditions. Furthermore, recently, it has been shown that small regulatory RNAs can silence nuclear-localized RNAs co-transcriptionally by the recruitment of nuclear RNAi defective (NRDE) factors that inhibit RNAP II during elongation.Citation62 It is possible that these small regulatory RNAs not only generate prematurely terminated transcripts by inhibition of RNAP II elongation but also, as discussed later in this review, they silence those erroneous transcripts by activation of a deadenylation process.
Cytoplasmic Polyadenylation/Deadenylation
In addition to its role in nuclear mRNA decay, the polyadenylation/deadenylation machineries have important roles in regulation of cytoplasmic mRNA stability, which is part of mRNA surveillance and cell differentiation processes. Deadenylation during cytoplamic mRNA degradation is biphasic.Citation63 First, the poly(A) tail is slowly shortened by the PAN2-PAN3 deadenylase complex to ∼100 nt; then the remaining poly(A) tail is rapidly degraded by the CCR4-CAF1 deadenylase complex to 8–12 nt.Citation64 The last phase of deadenylation is coincident with decapping and 5′-to-3′ decay. Antiproliferative factors from the B-cell translocation gene (BTG)/TOB family and the translation termination factor eRF3 have been described as general activators of mRNA deadenylation.Citation65–Citation67 While BTG2 function requires a direct interaction between BTG2 and POP2/CAF1 and CCR4 deadenylases,Citation67 TOB function requires the interaction between TOB and CCR4-CAF1 and PABPC1.Citation65 As BTG2 is involved in p53-dependent and p53-independent DDR, it may represent an alternative mechanism of global regulation of gene expression after specific stresses. Increased deadenylation rates are exhibited by mRNAs that are subject to NMD, to ARE- and/or miRNA-mediated decay.Citation68,Citation69 Besides, cytoplasmic polyadenylation has been described in germ-cell development, during early embryogenesis, in post-synaptic sites of nerve cells during learning and memory processes.Citation19,Citation20,Citation22,Citation23 These polyadenylation events lengthen the poly(A) tail of an mRNA with a shortened poly(A) tail, so that the mRNA will be translated. These shortened poly(A) tails are often less than 20 nucleotides, and are lengthened to around 80–150 nucleotides.
Gene expression in early animal development is regulated in large part by cytoplasmic polyadenylation and deadenylation (). Nuclear pre-mRNAs containing specific cis-acting signals in their 3′UTRs have long poly(A) tails, but once they are transported to the cytoplasm, their tails are shortened and those mRNAs become dormant. Once meiosis continues, the poly(A) tails are lengthened and mRNAs are translated. In fact, the sequential waves of polyadenylation and deadenylation drive the meiotic progression forward, preventing the back and forth slipping of the cell cycle phases in oocytes.Citation70,Citation71 In the early mouse embryo, cytoplasmic polyadenylation of maternal RNAs from the egg cell allows the cell to survive and grow even though transcription does not start until later. Polyadenylation of dormant mRNAs is controlled by the cytoplasmic polyadenylation element (CPE) and the hexamer AAUAAA that are the binding platforms for CPE-binding protein (CPEB) and CPSF, respectively. These small sequences present in the 3′UTR of target mRNAs were first described in Xenopus oocytes. Symplekin, which is also a component of the nuclear polyadenylation machinery, is involved in cytoplasmic polyadenylation by binding CPEB and CPSF and serving as a scaffold to bind the poly(A) polymerase Gld2.Citation23,Citation72,Citation73 PARN deadenylase is another important factor in this pathway.Citation73,Citation74 Interestingly, both Gld2 poly(A) polymerase and PARN deadenylase are part of a CPEB-containing complex ().Citation73 The shortening of the poly(A) tail could be explained by two alternative ways either the catalytic activities are modulated (i.e., PARN inhibits Gld2 activity and/or Gld2 activates PARN) or deadenylation by PARN is intrinsically more efficient and therefore out-competes polyadenylation by Gld2. Later, when oocytes mature, PARN is lost from the complex due to CPEB phosphorylation by the kinase Aurora A, allowing Gld2 to elongate the poly(A). The newly added poly(A) tail is protected from degradation by embryonic poly(A)-binding protein (ePAB), which first transiently associates with CPEB and then binds the poly(A) tail.Citation75 ePAB dissociation from CPEB is regulated by RINGO/cdk1, which is another protein kinase that phosphorylates CPEB.Citation76,Citation77
The translation of pre-existing maternal mRNAs occurs in a strict temporal order during meiotic cell cycle progression in Xenopus oocytes.Citation23 Cell cycle dependent signals that regulate the timing of cytoplasmic deadenylation are present in the 3′UTR. For example, polyadenylation response elements (PRE) and its binding factor Musashi, together with CPEB, contribute to the polyadenylation of early mRNAs.Citation78,Citation79 The phosphorylation of CPEB at multiple sites, including Ser174, is also very important for the activation of polyadenylation.Citation80–Citation83 Interestingly, late cytoplasmic polyadenylation also depends on CPEB phosphorylation by activated cdk1, which phosphorylates free CPEB labeling the protein for proteasomal degradation.Citation84–Citation86 CPEB associated with CPSF and symplekin is protected from phosphorylation and degradation. Thus, mRNAs that undergo late polyadenylation often have a CPE overlapping with the poly(A) signal that facilitate the interaction of CPEB with the other factors and the recognition of the CPE, avoiding CPEB phosphorylation and degradation and allowing polyadenylation at later stages.Citation84,Citation85,Citation87,Citation88 The initial deadenylation process is also highly regulated by different protein factors and cis-acting elements. Although, as mentioned before, the CPEB-PARN complex mediates this process (), other deadenylation factors, such as Pumilio ()Citation89–Citation91 and embryonic deadenylation element (EDEN)-binding protein (BP, )Citation92 might contribute to the regulation of the deadenylation of CPE-containing mRNAs. Some of the CPE-containing mRNAs also contain binding sites for Pumilio and EDEN-BP, indicating that the recruitment of these factors might be by both protein-protein and RNA-protein associations. Interestingly, Pumilio can bind not only CPEB but also the conserved deadenylase complex CCR4-POP2-NOT,Citation93 suggesting that deadenylases other than PARN might be involved in cytoplasmic deadenylation during development. Interestingly, the expression levels of many of the factors involved in cytoplasmic polyadenylation, such as cdk1, Aurora A, Gld2, are regulated by this process.Citation94,Citation95
Polyadenylation/deadenylation processes have also been described as part of the molecular mechanisms that allow long-term memory. While short-term memory is controlled by local activation of a cascade of kinases and phosphatases; long-term plasticity requires translational control of localized mRNAs. In mammals, dendritic mRNAs are maintained in a repressed state and are activated upon repetitive stimulation. Several regulatory proteins required for translational control in early development are thought to be required for memory formation, suggesting similar molecular mechanisms. In the brain, cytoplasmic polyadenylation is active during learning and could play a role in long-term potentiation, which is the strengthening of the signal transmission from one nerve cell to another in response to nerve impulses and is important for learning and memory formation. CPEB and Gld2, which are also involved in the activation of dormant mRNAs, are required for the stable maintenance of synaptic facilitation in AplysiaCitation96,Citation97 and for poly(A) elongation in Drosophila, respectively.Citation98 CPEB has also been shown to be important for neuronal synaptic plasticity, learning and memory using mouse knockout studies.Citation99,Citation100 Similar studies also show that CPEB plays a role in germ cell differentiation and synaptonemal complex formation.Citation101 Although several CPE-containing neuronal RNAs have been shown to undergo CPEB-dependent polyadenylation,Citation102 very few studies have validated the role of polyadenylation regulation of those RNAs in plasticity. Recently, the mRNA targets of the Drosophila neuronal CPEB, Orb2, have been identified.Citation103 Some of these targets include genes involved in neuronal growth, synapse formation and protein turnover, raising the possibility that Orb2-mediated polyadenylation might create a permissive environment for synaptic growth only at the activated synapse. It has also been shown that c-jun mRNA translation is regulated by CPEB and that growth hormone (GH) transcription via c-jun could mediate synaptic plasticity in the mouse hippocampus.Citation104 Interestingly, ARE-containing mRNAs are regulated by a polyadenylation/deadenylation mechanism both in the cytoplasm and in the nucleus.Citation49,Citation104 It has been shown that CPEB possesses a self-perpetuating prion-like behavior in invertebrates, and that the aggregated form of CPEB remains activated for a long period of time upregulating cytoplasmic polyadenylation and translation locally in dendrites.Citation105–Citation107 Further studies are required to determine if there is a functional connection between CPEB prion-like structure and its translational regulatory functions in different organisms.
In addition to these cytoplasmic processes, the polyadenylation/deadenylation machinery has an important role in the decay of mRNAs encoding premature translation termination codon (PTC). mRNAs with PTC are degraded in a pathway known as NMD.Citation108–Citation110 In this pathway, the nonsense transcripts are degraded either by deadenylation followed by 3′→5′ exonucleolytic degradation or by decapping without deadenylation.Citation108–Citation113 The molecular connections between NMD and deadenylation factors are still unclear. It has been shown that NMD factors RNA helicases Upf1, Upf2 and Upf3X can form a complex with the deadenylase PARN and activate deadenylation.Citation110 By contrast, a separate study reported no effect on NMD of PARN overexpression or knockdown, whereas inhibition of NMD was observed upon knockdown or upon overexpression of inactive mutants of the CCR4-CAF1 and PAN2-PAN3 deadenylase complexes.Citation63 On the other hand, another study has shown that the major deadenylase CCR4, which is involved in the turnover of normal mRNAs, is not involved in NMD.Citation114
Cis-Elements Involved in Polyadenylation/Deadenylation
Polyadenylation, deadenylation, mRNA stability and translation are often under the control of cis-acting regulatory elements within the 3′UTRs of eukaryotic mRNAs. These sequence elements recruit trans-acting protein factors that regulate those processes and affect gene expression. Examples of these cis-acting elements include APA signals, AREs and miRNA target sites. The use of different APA signals generates a diversity of mRNA isoforms that carry different arrangements of AREs and miRNA target sites. Both sequence and location of these elements are responsible for their accurate regulatory functions. Many studies support the idea that miRNAs destabilize mRNA through deadenylation pathways, while AREs function in mRNA stability by either preventing mRNA from degradation by exosome or by recruiting the exosome to decrease the mRNA stability. The degradation of these mRNAs is linked to the appearance of processing bodies (p-bodies), which are cytoplasmic foci involved in mRNA turnover. One central component of the p-bodies, Pat1b, has been shown to physically connect deadenylation with decapping by the recruitment of both the CCR4-CAF1-NOT deadenylation complex and the Dcp1-Dcp2 decapping complex.Citation115
About 54% of human genes have more than one conserved polyadenylation sites that show different efficiencies of polyadenylation ().Citation116 Several cis-acting elements within the 3′UTR are responsible for the selection among these APA sites. The hexamer AAUAAA and U/GU-rich elements are the core mammalian mRNA polyadenylation elements that bind CPSF and CstF, respectively; and the cleavage site lies in between these two core elements. Additional cis-acting elements that function to alter the efficiency of polyadenylation have also been identified as auxiliary RNA elements.Citation117,Citation118 However, once the signal is recognized by the cleavage/polyadenylation complex in the nucleus, the full-length poly(A) tail is added to the cleaved 3′ end. No regulatory mechanisms have been described yet at this first round of polyadenylation, suggesting that addition of a poly(A) tail is a default modification after mRNA 3′ cleavage in the nucleus. Although a direct connection between APA and the polyadenylation/deadenylation machinery has not been described, the selection between the distal or proximal alternative signals might cause the inclusion or exclusion of other cis-acting RNA elements involved in polyadenylation/deadenylation processes. The relevance of these regulatory processes is highlighted by the finding of changes in the length of the 3′UTRs of different mRNAs, which change the number of miRNA target sites and AREs, in cancer cellsCitation10,Citation11 and during cell differentiation.Citation12–Citation14
miRNAs comprise a large family of small single-stranded non-coding RNAs (∼21 nts in length), which are encoded within the genome of species ranging from protozoans to plants to mammals. In mammals, it is predicted that the regulation of more than 60% of all protein-coding genes are mediated by miRNAs via their imperfectly base-pairing with the target mRNA 3′UTR through their 5′-proximal “seed” region.Citation119–Citation123 It has been shown that each mRNA could be regulated by more than one miRNA, and each miRNA could base-pair with more than one target mRNAs. miRNAs control protein synthesis either through translation regulation and/or through deadenylation activation, which leads subsequently to mRNA degradation ().Citation123–Citation126 These two mechanisms have been identified as two distinct independent pathways: the ability of miRNAs to expedite deadenylation does not depend on decreased translation; nor does translational repression by miRNAs require a poly(A) tail.Citation127
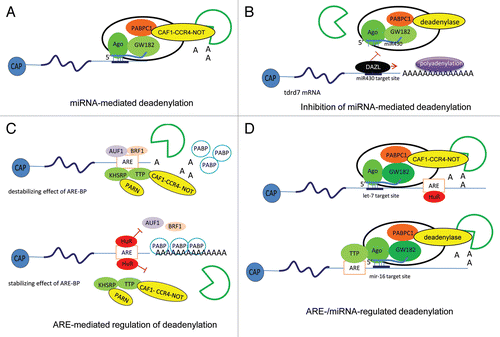
miRNAs function in the form of ribunucleoprotein complexes known as miRNA-induced silencing complexes (miRISCs), which deliver miRNAs to their mRNA targets. miRISCs facilitate degradation of miRNA targeting mRNAs via a biphasic deadenylation followed by Dcp1-Dcp2 complex-directed decapping, and subsequent exonucleolytic digestion.Citation128 Argonaute (Ago) family proteins are one of the best-characterized core components of miRISCs. Agos recruit another group of factors, the GW182 (TNRC6 in humans), which are crucial for accelerated deadenylation by miRNA.Citation129–Citation131 Interestingly, GW182 dissociates from miRISCs at a step of silencing downstream of deadenylation, indicating that GW182 is essential to initiate silencing by activating deadenylation but is not required to maintain silencing.Citation132,Citation133 The cytoplasmic polyadenylate binding protein 1 (PABPC1) is an additional protein component that is critical for the miRNA-mediated deadenylation. It has been shown that a conserved motif in GW182 interacts with the C-terminal domain of PABPC1 and that this interaction and the activity of PABPC1 contribute to miRNA-mediated poly(A) removal.Citation133–Citation135 One of the best studied deadenylases involved in miRISCs is the CAF1-CCR4-NOT1 complex, which contains two protein factors with deadenylase activity, CCR4 and CAF1. It has been shown that the CAF1-CCR4-NOT complex associates with PABPC1, and the deadenylase activity of the CAF1-CCR4-NOT complex is necessary for the miRNA-mediated deadenylation.Citation133 Supporting these results, both the knockdown of CAF1 or NOT1 expressionCitation132,Citation134 and the overexpression of CCR4 or CAF1 mutants could significantly reduce miRNA-mediated deadenylation and mRNA decay, but not translational repression.Citation136 Pan2-Pan3 deadenylase has also been described to play a role in miRNA-mediated deadenylation.Citation128 How and if PARN, one of the three major deadenylases in human cells, is involved in miRNA-mediated deadenylation still remains to be established. On the basis of these studies, it has been proposed that the miRNA-mediated degradation of mRNAs involves the association of Ago proteins to the miRNAs and the recruitment of GW182 to the target mRNAs via its N-terminal domain; then the GW182 C-terminal silencing domain recruits the deadenylase complex through the interaction with PABPC1. However, the deadenylase functions of CCR4-NOT can be reconstituted by direct tethering of miRISC to the mRNA, suggesting that the activation of deadenylation by miRISC is independent of miRNA base pairing.Citation128,Citation136 It has been shown in human cells and in primordial germ cells of zebrafish that some RNA-binding proteins, such as Dnd1 and DAZL, can protect certain mRNAs from miRNA-mediated repression by binding mRNAs and prohibiting miRNAs from associating with their target sites ().Citation137,Citation138 Interestingly, DAZL not only reverts the miRNA-mediated repression by miR-430 but also induces polyadenylation of the tdrd7 mRNA irrespective of the function of miRNA.Citation138
The role of miRNA-mediated deadenylation in different biological processes has been studied in different systems. For example, it has been shown that let-7 miRNPs directly activate deadenylation of target mRNAs in a cell-free system,Citation139 and that the overexpression of let-7 miRNAs decreases the mRNA levels of oncogenes, such as c-mycCitation140 and RAS.Citation141 These results indicate that a decrease in let-7 miRNA levels might lead to tumorigenicity in lung and colon cancers.Citation141,Citation142 The processing of let-7 precursors is blocked by lin-28B, a putative RNA-binding protein highly expressed in hepatocellular carcinoma.Citation143 The overexpression of lin-28B enhances tumorigenecity by increasing the expression of the oncogenic let-7 targets.Citation143 miR-125b has also been described to participate in miRNA-mediated deadenylation and in the reduction of cellular abundance of targeting mRNAs.Citation144 Interestingly, the overexpression of miR-125b correlates with the downregulation of lin-28.Citation145 These studies indicate that miRNAs might play crucial roles in multiple oncogenic pathways. In fact, it has been suggested that miRNAs might function as oncogenes and promote cancer development by negatively regulating tumor suppressor genes and/or genes that control cell differentiation of apoptosis.Citation146,Citation147
About 12% of mammalian mRNAs bear important regulatory signal AREs in their 3′UTRs, which have been shown to play significant roles in mRNA stability regulation.Citation148 The ARE typically contains one or several AUUUA pentamer repeats within a U-rich region of the 3′UTR.Citation149–Citation151 ARE sequences are frequently present in genes that encode tightly regulated proteins involved in cell growth regulation, cell differentiation and responses to external stimuli. The destabilizing functions of AREs are important because in their absence proto-oncogenes, such as c-fos, c-myc, c-jun, could become oncogenes.Citation152 Other mRNAs, such as IL-3, need AREs in order to inhibit the growth of autocrine tumor mast cells by an immunosuppressant cyclosporin A.Citation153 A number of trans-acting factors, known as ARE-binding proteins (ARE-BPs), regulate ARE-mediated decay by either inhibiting or activating deadenylation, and subsequently change the stability of ARE-containing mRNAs ().Citation154 The ARE-BPs that promote ARE-containing mRNAs decay include tristetraprolin (TTP), butyrate response factor 1 (BRF1), AU-rich binding factor 1 (AUF1) and KH-type splicing regulatory protein (KHSRP or KSRP). So far only Hu protein R (HuR) has been shown to play a role in stabilizing ARE-containing mRNAs.Citation154–Citation156 ARE-BPs regulate ARE-mediated decay of mRNAs by recruiting the deadenylases to the target mRNAs in different cellular conditions. For example, TTP directs its target ARE-containing mRNA tumor necrosis factor (α-TNF) for degradation by expediting removal of the poly(A) tail. Interestingly, the phosphorylation of TTP by mitogen-activated protein kinase-activated protein kinase 2 reduces the ability of TTP to promote deadenylation by inhibiting the recruitment of CAF1 deadenylase, indicating that the CAF1-CCR4-NOT deadenylase complex is responsible for TTP-directed deadenylation.Citation157–Citation160 Supporting the idea that CAF1 deadenylase plays critical role in ARE-mediated deadenylation, knockdown of CAF1 has been shown to abrogate the deadenylation and decay of the ARE-containing α-globin mRNA.Citation161,Citation162 Besides the CAF1-CCR4-NOT deadenylase complex the deadenylase PARN has also been suggested to be involved in ARE-mediated deadenylation and to promote TTP-directed deadenylation in vitro.Citation52,Citation163,Citation164 KSRP recruits PARN to ARE-containing mRNAs to initiate the poly(A) tail shortening that precedes the degradation by the exosome.Citation165 On the other hand, ARE-BPs, like HuR, can stabilize ARE-containing mRNAs by blocking the recruitment of deadenylases or the exosome.Citation166
Interestingly, some of the seed signals recognized by miRNA overlap with AREs in the 3′UTR. Although the exact contribution of miRNAs, miRISC, AREs and ARE-BPs to mRNA decay has not been elucidated yet, recent studies have described a functional overlap between AREs- and microRNAs-mediated mRNA turnover pathways (). It has been described that miRNAs can functionally interact with ARE-BPs, and that Dicer and Ago are required for ARE-mediated decay.Citation167 For example, it has been shown that HuR can bind AREs present in c-myc 3′UTR at a site proximal to that recognized by let-7 miRNA. HuR appeared to facilitate the targeting of let-7-loaded miRISC to an adjacent region of HuR binding site, and to mediate the reduction of c-Myc mRNA levels.Citation168 Another example is the cooperation of ARE-BP TTP and miR-16 in targeting tumor necrosis factor-α mRNA for ARE-mediated mRNA degradation.Citation167 TTP does not bind directly miR-16 but it forms a complex with miRISC, and that complex recruits the deadenylase and the exosome for mRNA degradation. HuR can also relieve CAT-1 mRNA from miR-122 repression upon stress in human liver cells.Citation169
Conclusions
Understanding the mechanisms and consequences of the changes in the lengths of mRNA poly(A) tails has been a matter of investigation for many years and it has been established that modulation of the length of the poly(A) tail of an mRNA is a widespread strategy used to control protein production and mRNA stability. This mechanism of post-transcriptional regulation has turned out to be an essential mechanism for proper control of gene expression in different cellular conditions. The potential for mRNA stability and translation control is enormous encompassing hundreds of protein and miRNA regulators, a multitude of deadenylases and abundant cis-acting signals in the 3′UTR. The data reviewed here clearly indicate that poly(A) tail length results from a balance between concomitant deadenylation and polyadenylation in numerous cellular responses, such as nuclear mRNA surveillance, DDR, development and long-term memory. Despite intensive research and substantial progress during recent years, we still do not know in detail the underlying molecular mechanism behind polyadenylation/deadenylation in these processes and how the two processes are interconnected to each other. Thus, several fundamental questions are still waiting to be solved. For example: does a homologue to the TRAMP complex exist in mammalian cells? Does the CstF/PARN complex play a role in processes other than DDR? What are the mRNA targets regulated by CstF/PARN? Do other cis-acting sequences regulate cytoplasmic polyadenylation? Are those sequences present in different group of mRNAs? Which are the PAPs and deadenylases involved in each of these processes? What are the direct interactions between protein factors and RNA elements? Is the polyadenylation/deadenylation machinery involved in NMD? Is polyadenylation also involved in the miRNA-dependent regulation of mRNA stability and translational repression? As growing evidence reveals connections between 3′ end mRNA processing events and human diseases,Citation170 what is the medical relevance of the regulation of polyadenylation/deadenylation machineries? The development of novel approaches for studying 3′ end processing and the determination of new protein factors and cis-elements involved in these reactions will be essential to address some of these basic questions.
Abbreviations
| RNAP II | = | RNA polymerase II |
| CPSF | = | cleavage-polyadenylation specificity factor |
| CstF | = | cleavage stimulation factor |
| PAP | = | poly(A) polymerase |
| PABNI | = | poly(A)-binding protein nuclear I |
| 3′UTR | = | 3′ untranslated region |
| PARN | = | poly(A)-specific ribonuclease |
| PAN | = | poly(A) nuclease |
| DDR | = | DNA damage response |
| NMD | = | non-sense mediated decay |
| miRNA | = | microRNA |
| ARE | = | AU-rich element |
| CBP80 | = | cap binding protein 80 |
| CPE | = | cytoplasmic polyadenylation element |
| CPEB | = | CPE-binding protein |
| ePAB | = | embryonic poly(A)-binding protein |
| EDEN | = | embryonic deadenylation element |
| PTC | = | premature translation termination codon |
| APA | = | alternative polyadenylation |
| p-body | = | processing body |
| miRISC | = | miRNA-induced silencing complex |
| Ago | = | argonaute |
| PABPC1 | = | polyadenylate binding protein cytoplasmic 1 |
| ARE-BP | = | ARE-binding protein |
| TTP | = | tristetraprolin |
| BRF1 | = | butyrate response factor 1 |
| AUF1 | = | AU-rich binding factor 1 |
| KHSRP or KSRP | = | KH-type splicing regulatory protein |
| α-TNF | = | tumor necrosis factor |
| NRDE | = | nuclear RNAi defective |
| Dun1 | = | DNA damage response protein kinase DUN1 |
Figures and Tables
Figure 1 Model for nuclear polyadenylation/deadenylation machineries involved in RNA surveillance and DDR. (A) In yeast, the TRAMP complex-mediated polyadenylation/deadenylation is mechanistically implicated in mRNA surveillance. The TRAMP complex and the exosome are recruited to defective RNAs at sites of active transcription by RNAP II. The TRAMP complex, which contains Trf4p and Trf5p (noncanonical PAPs), Mtr4p (RNA helicase) and Air1p/Air2p (RNA binding protein), adds a ∼40 nucleotides tail to the 3′ end of target RNA to mark it for degradation by the exosome. The CCR4-NOT deadenylase can interact physically and functionally with both the exosome and the TRAMP complex. Rrp6, a nuclear exosome component, is necessary not only for RNA exonucleolytic degradation but also for transcription termination. (B) In mammalian cells, the nuclear polyadenylation/deadenylation switch regulates RNA turnover in response to DNA damage. In the absence of DNA damage treatment, CBP80 binds to nuclear deadenylase PARN and inhibits its deadenylase activity. The elongating RNAP II-CstF holoenzyme is active and polyadenylation takes place generating normal levels of mRNA. Upon UV-induced DNA damage, PARN dissociates from CBP80 and interacts with the CstF1/BARD1 complex. Interestingly, the formation of PARN/CstF1/BARD1 complex not only inhibits RNA 3′ end cleavage, but at the same time also activates the deadenylation by PARN and leads to RNA degradation.
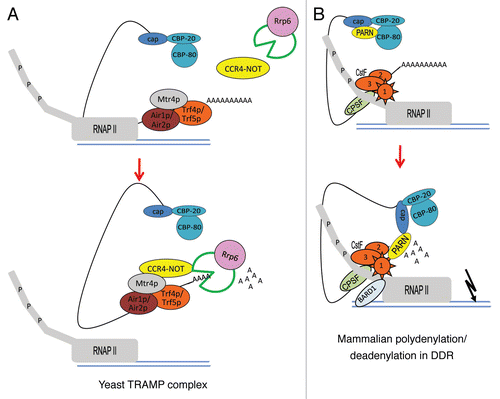
Figure 2 Model for cytoplasmic polyadenylation/deadenylation during cell differentiation processes. (A) The deadenylation and polyadenylation of dormant mRNAs are controlled by cis-elements at 3′UTR, such as the CPE and the AAUAAA signals, which are recognized by the polyadenylation factors CPEB and CPSF, respectively. Poly(A) polymerase Gld2 and deadenylase PARN are crucial components of CPEB-containing complexes in the regulation of deadenylation/polyadenylation. After nuclear polyadenylated mRNAs are transported to the cytoplasm, their poly(A) tails are shortened by PARN deadenylase associated to CPEB-containing complexes. Although Gld2 polymerase is also associated to CPEB-containing complexes, its activity is inhibited. During meiotic cell cycle progression, CPEB is phosphorylated and PARN dissociates from CPEB-containing complex, allowing Gld2 to elongate the poly(A) tail and to label the CPE containing mRNAs for translation. (B) Model for translation repression by deadenylation. As deadenylation is the first step in this highly regulated process of cytoplasmic polyadenylation/deadenylation during meiosis, other deadenylation factors are also involved allowing a strict spatial and temporal regulation. Many CPE containing mRNAs also contain other signals in their 3′UTR, such as Pumilio and/or embryonic deadenylation element (EDEN) binding sites, which are recognized by the deadenylation factors Pumilio and EDEN-BP, respectively. While Pumilio interacts with the conserved deadenylase complex CCR4-POP2-NOT (shown to the left), EDEN-BP interacts with PARN to deadenylate mRNAs (shown to the right). Some of these cis-acting RNA sequences are present in the 3′UTR of the same mRNAs, allowing numerous alternative mechanisms to regulate deadenylation/polyadenylation of mRNAs.
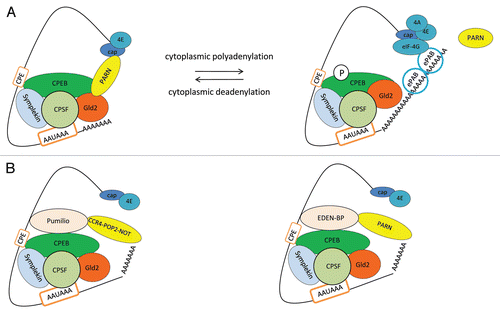
Figure 3 Schematic diagrams of cis-acting RNA sequences at the 3′UTR involved in polyadenylation/deadenylation processes. The selection between the proximal or distal alternative polyadenylation signals leads to the exclusion or inclusion of cis-acting RNA sequences, such as miRNA target sites and ARE, that could mediate polyadenylation/deadenylation processes.
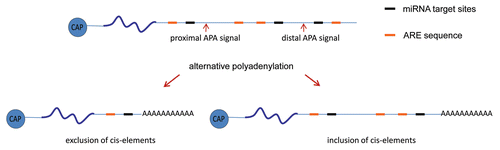
Figure 4 Models of multicomponent complexes required for regulation of polyadenylation/deadenylation processes. (A) miRNA-mediated deadenylation. miRISCs, which contain Ago, GW182, PABPC1 and either CAF1-CCR4-NOT (as indicated) or Pan2-Pan3 (not shown) deadenylases, deliver miRNAs to the target mRNAs and mediate deadenylation, which leads subsequently to mRNA degradation. (B) Inhibition of miRNA-mediated deadenylation. The RNA-binding protein DAZL overcomes the inhibitory effect of miRNA by inducing polyadenylation and leading to active translation even in the presence of miRNA. (C) ARE-mediated regulation of deadenylation. The ARE-binding proteins (BP) mediate destabilization and/or stabilization of the ARE-containing mRNAs. ARE-BPs, such as AUF1, TTP, BRF1 and KHSRP, recruit deadenylases, such as PARN and CAF1-CCR4-NOT, to target ARE-mRNAs and initiate the deadenylation process that precedes degradation. Another ARE-BP, HuR, plays a role in stabilizing ARE-containing mRNAs by blocking the binding of ARE-BPs involved in the destabilization of ARE-mRNAs, such as AUF1, TTP and KHSRP. This competition stabilizes the association of PABP to the poly(A) tail or prevents the recruitment of deadenylases to the ARE-mRNA. (D) ARE- and miRNA-regulated deadenylation. Cooperation of AREs-BPs, miRNAs, deadenylases and exosome is essential for the regulation of mRNA stability. The recruitment of the ARE-BPs HuR or TTP to the ARE sequence assists the targeting of let-7- or miR-16-loaded miRISC complexes, respectively, to the most proximal site to the ARE sequence.
Acknowledgments
We apologize to those authors whose work was not cited because of space limitations. The authors are supported by National Institute of General Medical Sciences grant SC1GM083806 to F.E.K. and the Swedish Research Council and the Linneus Support from the Swedish Research Council to the Uppsala RNA Research Centre to A.V.
References
- Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 2005; 17:251 - 256; PMID: 15901493
- Sheets MD, Wickens M. Two phases in the addition of a poly(A) tail. Genes Dev 1989; 3:1401 - 1412; PMID: 2575065
- Gilmartin GM. Eukaryotic mRNA 3′ processing: A common means to different ends. Genes Dev 2005; 19:2517 - 2521; PMID: 16264187
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation and interrelationships with other steps in mRNA synthesis. Mircrobiol Mol Biol Rev 1999; 63:405 - 445; PMID: 10357856
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 2008; 65:1099 - 1122; PMID: 18158581
- Kühn U, Gündel M, Knoth A, Kerwitz Y, Rüdel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem 2009; 284:22803 - 22814; PMID: 19509282
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 2009; 33:365 - 376; PMID: 19217410
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol 2008; 20:260 - 265; PMID: 18436438
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 2010; 38:2757 - 2774; PMID: 20044349
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009; 138:673 - 684; PMID: 19703394
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, et al. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Can Res 2009; 69:9422 - 9430; PMID: 19934316
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008; 320:1643 - 1647; PMID: 18566288
- Zlotorynski E, Agami R. A PASport to cellular proliferation. Cell 2008; 134:208 - 210; PMID: 18662535
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA 2009; 106:7028 - 7033; PMID: 19372383
- Huang Y, Carmichael GG. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol 1996; 16:1534 - 1542; PMID: 8657127
- Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA 1998; 4:1321 - 1331; PMID: 9814754
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 2004; 11:121 - 127; PMID: 14749774
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 2008; 9:337 - 344; PMID: 18334997
- Colegrove-Otero LJ, Minshall N, Standart N. RNA-binding proteins in early development. Crit Rev Biochem Mol Biol 2005; 40:21 - 73; PMID: 15804624
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 2007; 8:776 - 789; PMID: 17848965
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 2007; 8:113 - 126; PMID: 17245413
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol 2007; 23:405 - 433; PMID: 17506698
- Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta 2008; 1779:217 - 229; PMID: 18316045
- Cevher MA, Kleiman FE. Connections between 3′-end processing and DNA damage response. Wiley Interdisciplinary Reviews-RNA 2010; 1:193 - 199
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, et al. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 2002; 21:165 - 174; PMID: 11782436
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 2006; 7:529 - 539; PMID: 16829983
- Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep 2007; 8:651 - 657; PMID: 17603538
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 2000; 102:765 - 775; PMID: 11030620
- Burkard KT, Butler JS. A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 2000; 20:604 - 616; PMID: 10611239
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 2001; 413:538 - 542; PMID: 11586364
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3′-end quality control and nuclear degradation. Mol Cell Biol 2002; 22:8254 - 8266; PMID: 12417728
- Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol Cell 2002; 9:1285 - 1296; PMID: 12086625
- Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol 2003; 23:5502 - 5515; PMID: 12897126
- Jensen TH, Rosbash M. Co-transcriptional monitoring of mRNP formation. Nat Struct Biol 2003; 10:10 - 12; PMID: 12490887
- Jensen TH, Boulay J, Olesen JR, Colin J, Weyler M, Libri D. Modulation of transcription affects mRNP quality. Mol Cell 2004; 16:235 - 244; PMID: 15494310
- Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol Cell Biol 2004; 24:9646 - 9657; PMID: 15485930
- Fasken MB, Corbett AH. Process or perish: quality control in mRNA biogenesis. Nat Struct Mol Biol 2005; 12:482 - 488; PMID: 15933735
- Kuai L, Das B, Sherman F. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 2005; 102:13962 - 13967; PMID: 16166263
- Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol Cell Biol 2005; 25:9996 - 10004; PMID: 16260613
- Wang SW, Stevenson AL, Kearsey SE, Watt S, Bähler J. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol 2008; 28:656 - 665; PMID: 18025105
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell 2006; 23:853 - 864; PMID: 16973437
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 2005; 121:725 - 737; PMID: 15935759
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005; 121:713 - 724; PMID: 15935758
- Azzouz N, Panasenko OO, Colau G, Collart MA. The CCR4-NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS One 2009; 4:6760; PMID: 19707589
- Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem 2010; 285:3540 - 3547; PMID: 19955569
- Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev 2004; 18:2652 - 2662; PMID: 15489286
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell 2006; 24:943 - 953; PMID: 17189195
- Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci USA 2007; 104:10412 - 10417; PMID: 17563387
- Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, et al. Nuclear deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. EMBO J 2010; 29:1674 - 1687; PMID: 20379136
- Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3′-end formation to DNA damage and tumor suppression. Cell 2001; 104:743 - 753; PMID: 11257228
- Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, Manley JL. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev 2005; 19:1227 - 1237; PMID: 15905410
- Balatsos NA, Nilsson P, Mazza C, Cusack S, Virtanen A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC). J Biol Chem 2006; 281:4517 - 4522; PMID: 16317009
- Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol 2003; 23:3798 - 3812; PMID: 12748283
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 2006; 12:1084 - 1091; PMID: 16601207
- Blattner C, Kannouche P, Litfin M, Bender K, Rahmsdorf HJ, Angulo JF, Herrlich P. UV-Induced stabilization of c-fos and other short-lived mRNAs. Mol Cell Biol 2000; 20:3616 - 3625; PMID: 10779351
- Bollig F, Winzen R, Kracht M, Ghebremedhin B, Ritter B, Wilhelm A, et al. Evidence for general stabilization of mRNAs in response to UV light. Eur J Biochem 2002; 26:5830 - 5839; PMID: 12444971
- Gowrishankar G, Winzen R, Bolling F, Ghebremedhin B, Redich N, Ritter B, et al. Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biol Chem 2005; 386:1287 - 1293; PMID: 16336123
- Hammet A, Pike BL, Heierhorst J. Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A)-nuclease complex contributes to survival of replication blocks. J Biol Chem 2002; 277:22469 - 22474; PMID: 11953437
- Traven A, Hammet A, Tenis N, Denis CL, Heierhorst J. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics 2005; 169:65 - 75; PMID: 15466434
- Westmoreland TJ, Marks JR, Olson JA Jr, Thompson EM, Resnick MA, Bennett CB. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae. Eukaryotic Cell 2004; 3:430 - 446; PMID: 15075273
- Gaillard H, Wellinger RE, Aguilera A. Methods to study transcription-coupled repair in chromatin. Methods Mol Biol 2009; 523:141 - 159; PMID: 19381941
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 2010; 465:1097 - 1102; PMID: 20543824
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 2005; 12:1054 - 1063; PMID: 16284618
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev 1993; 7:1632 - 1643; PMID: 8393418
- Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, et al. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 2007; 27:7791 - 7801; PMID: 17785442
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, et al. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 2007; 21:3135 - 3148; PMID: 18056425
- Mauxion F, Faux C, Séraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J 2008; 27:1039 - 1048; PMID: 18337750
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 2005; 19:351 - 361; PMID: 15687258
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006; 312:75 - 79; PMID: 16484454
- Xiong W, Ferrell JE. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature 2003; 426:460 - 465; PMID: 14647386
- Belloc E, Piqué M, Méndez R. Sequential waves of polyadenylation and deadenylation define a translation circuit that drives meiotic progression. Biochem Soc Trans 2008; 36:665 - 670; PMID: 18631137
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 2004; 119:641 - 651; PMID: 15550246
- Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 2006; 24:173 - 183; PMID: 17052452
- Körner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J 1998; 17:5427 - 5437; PMID: 9736620
- Kim JH, Richter JD. Measuring CPEB-mediated cytoplasmic polyadenylation-deadenylation in Xenopus laevis oocytes and egg extracts. Methods Enzymol 2008; 448:119 - 138; PMID: 19111174
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev 2001; 15:774 - 788; PMID: 11274061
- Wilkie GS, Gautier P, Lawson D, Gray NK. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol Cell Biol 2005; 25:2060 - 2071; PMID: 15713657
- Charlesworth A, Cox LL, MacNicol AM. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J Biol Chem 2004; 279:17650 - 17659; PMID: 14752101
- Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J 2006; 25:2792 - 2801; PMID: 16763568
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell 2000; 6:1253 - 1259; PMID: 11106762
- Ma C, Cummings C, Liu XJ. Biphasic activation of Aurora-A kinase during the meiosis I-meiosis II transition in Xenopus oocytes. Mol Cell Biol 2003; 23:1703 - 1716; PMID: 12588989
- Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev 2007; 21:2571 - 2579; PMID: 17938241
- Keady BT, Kuo P, Martínez SE, Yuan L, Hake LE. MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J Cell Sci 2007; 120:1093 - 1103; PMID: 17344432
- Ballantyne S, Daniel DL Jr, Wickens M. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol Biol Cell 1997; 8:1633 - 1648; PMID: 9285830
- De Moor CH, Richter JD. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol Cell Biol 1997; 17:6419 - 6426; PMID: 9343404
- Charlesworth A, Ridge JA, King LA, MacNicol MC, MacNicol AM. A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J 2002; 21:2798 - 2806; PMID: 12032092
- Mendez R, Barnard D, Richter JD. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J 2002; 21:1833 - 1844; PMID: 11927567
- Setoyama D, Yamashita M, Sagata N. Mechanism of degradation of CPEB during Xenopus oocyte maturation. Proc Natl Acad Sci USA 2007; 104:18001 - 18006; PMID: 17986610
- Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2 and a cytoplasmic polyadenylation element-binding protein. J Biol Chem 2001; 276:20945 - 20953; PMID: 11283000
- Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev 2006; 20:199 - 209; PMID: 16418484
- Piqué M, López JM, Foissac S, Guigó R, Méndez R. A combinatorial code for CPE-mediated translational control. Cell 2008; 132:434 - 448; PMID: 18267074
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J 1998; 17:278 - 287; PMID: 9427761
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 2002; 21:1427 - 1436; PMID: 11889048
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′ untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev 1994; 8:926 - 938; PMID: 7926777
- Rouhana L, Wickens M. Autoregulation of GLD-2 cytoplasmic poly(A) polymerase. RNA 2007; 13:188 - 199; PMID: 17164476
- Liu J, Schwartz JH. The cytoplasmic polyadenylation element binding protein and polyadenylation of messenger RNA in Aplysia neurons. Brain Res 2003; 959:68 - 76; PMID: 12480159
- Thom G, Minshall N, Git A, Argasinska J, Standart N. Role of cdc2 kinase phosphorylation and conserved N-terminal proteolysis motifs in cytoplasmic polyadenylation-element-binding protein (CPEB) complex dissociation and degradation. Biochem J 2003; 370:91 - 100; PMID: 12401129
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proc Natl Acad Sci USA 2008; 105:14644 - 14649; PMID: 18780789
- Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem 2006; 13:4 - 7; PMID: 16452649
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol 2006; 16:102 - 110; PMID: 16418001
- Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell 2001; 1:201 - 213; PMID: 11702780
- Du L, Richter JD. Activity-dependent polyadenylation in neurons. RNA 2005; 11:1340 - 1347; PMID: 16043499
- Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation and protein turnover. Proc Natl Acad Sci USA 2010; 107:11987 - 11992; PMID: 20547833
- Zearfoss NR, Alarcon JM, Trifilieff P, Kandel E, Richter JD. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J Neurosci 2008; 28:8502 - 8509; PMID: 18716208
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 2003; 115:879 - 891; PMID: 14697205
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 2003; 115:893 - 904; PMID: 14697206
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron 2009; 61:10 - 26; PMID: 19146809
- Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol Cell 2003; 11:1405 - 1413; PMID: 12769863
- Takahashi S, Araki Y, Sakuno T, Katada T. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J 2003; 22:3951 - 3959; PMID: 12881429
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating and exonucleolytic activities. Mol Cell 2003; 12:675 - 687; PMID: 14527413
- Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 2003; 23:4805 - 4813; PMID: 12832468
- Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta 2008; 1779:538 - 549; PMID: 18657639
- Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 2001; 106:607 - 617; PMID: 11551508
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 2002; 295:2258 - 2261; PMID: 11910109
- Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol 2010; 30:4308 - 4323; PMID: 20584987
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 2005; 33:201 - 212; PMID: 15647503
- Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′UTR. RNA 2007; 13:1103 - 1115; PMID: 17507659
- Lutz CS. Alternative polyadenylation: A twist on mRNA 3′ end formation. ACS Chem Biol 2008; 3:609 - 617; PMID: 18817380
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol 2007; 23:175 - 205; PMID: 17506695
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215 - 233; PMID: 19167326
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet 2009; 10:94 - 108; PMID: 19148191
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19:92 - 105; PMID: 18955434
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?. Nat Rev Genet 2008; 9:102 - 114; PMID: 18197166
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132:9 - 14; PMID: 18191211
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA 2009; 15:21 - 32; PMID: 19029310
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 2009; 21:452 - 460; PMID: 19450959
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 2006; 103:4034 - 4039; PMID: 16495412
- Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol 2009; 16:1160 - 1166; PMID: 19838187
- Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 2009; 15:1059 - 1066; PMID: 19383768
- Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA 2009; 15:1078 - 1089; PMID: 19398495
- Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA 2009; 15:781 - 793; PMID: 19304925
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 2006; 20:1885 - 1898; PMID: 16815998
- Zekri L, Huntzinger E, Heimstädt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol 2009; 29:6220 - 6231; PMID: 19797087
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 2009; 35:868 - 880; PMID: 19716330
- Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol 2010; 17:238 - 240; PMID: 20098421
- Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol 2010; 30:1486 - 1494; PMID: 20065043
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007; 131:1273 - 1286; PMID: 18155131
- Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One 2009; 4:7513; PMID: 19838299
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev 2007; 21:1857 - 1862; PMID: 17671087
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a downregulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007; 67:9762 - 9770; PMID: 17942906
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell 2005; 120:635 - 647; PMID: 15766527
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 2006; 29:903 - 906; PMID: 16651716
- Wang YC, Chen YL, Yuan RH, Pan HW, Yang WC, Hsu HC, et al. Lin-28B expression promotes transformation and invasion in human hepatocellular carcinoma. Carcinogenesis 2010; 31:1516 - 1522; PMID: 20525879
- Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol 2005; 25:9198 - 9208; PMID: 16227573
- Eda A, Tamura Y, Yoshida M, Hohjoh H. Systematic gene regulation involving miRNAs during neuronal differentiation of mouse P19 embryonic carcinoma cell. Biochem Biophys Res Commun 2009; 388:648 - 653; PMID: 19679099
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302:1 - 12; PMID: 16989803
- Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: A primer. Am J Pathol 2007; 171:728 - 738; PMID: 17724137
- Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene 2001; 265:11 - 23; PMID: 11255003
- Lagnado CA, Brown CY, Goodall GJ. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUA UUU A(U/A)(U/A). Mol Cell Biol 1994; 14:7984 - 7995; PMID: 7969138
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 1995; 20:465 - 470; PMID: 8578590
- Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUA UUU AUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol 1995; 15:2219 - 2230; PMID: 7891716
- Schiavi SC, Belasco JG, Greenberg ME. Regulation of proto-oncogene mRNA stability. Biochim Biophys Acta 1992; 1114:95 - 106; PMID: 1457466
- Nair AP, Hahn S, Banholzer R, Hirsch HH, Moroni C. Cyclosporin A inhibits growth of autocrine tumor cell lines by destabilizing interleukin-3 mRNA. Nature 1994; 369:239 - 242; PMID: 8183344
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles?. Nucleic Acids Res 2006; 33:7138 - 7150; PMID: 16391004
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem 1996; 271:8144 - 8151; PMID: 8626503
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 1998; 17:3448 - 3460; PMID: 9628880
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 1999; 19:4311 - 4323; PMID: 10330172
- Dean JL, Sully G, Clark AR, Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilization. Cell Signal 2004; 16:1113 - 1121; PMID: 15240006
- Winzen R, Gowrishankar G, Bollig F, Redich N, Resch K, Holtmann H. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol Cell Biol 2004; 24:4835 - 4847; PMID: 15143177
- Marchese FP, Aubareda A, Tudor C, Saklatvala J, Clark AR, Dean JL. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem 2010; 285:27590 - 27600; PMID: 20595389
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 2008; 182:89 - 101; PMID: 18625844
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res 2008; 36:3374 - 3388; PMID: 18442996
- Körner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem 1997; 272:10448 - 10456; PMID: 9099687
- Lin WJ, Duffy A, Chen CY. Localization of AU-rich element-containing mRNA in cytoplasmic granules containing exosome subunits. J Biol Chem 2007; 282:19958 - 19968; PMID: 17470429
- Gherzi R, Lee KY, Briata P, Wegmüller D, Moroni C, Karin M, et al. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell 2004; 14:571 - 583; PMID: 15175153
- Westmark CJ, Bartleson VB, Malter JS. RhoB mRNA is stabilized by HuR after UV light. Oncogene 2005; 24:502 - 511; PMID: 15543229
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005; 120:623 - 634; PMID: 15766526
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 2009; 23:1743 - 1748; PMID: 19574298
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006; 125:1036 - 1038; PMID: 16777601
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 2008; 27:482 - 498; PMID: 18256699