Abstract
Together, circulating BAFF and dominant receptor BAFF-R homeostatically regulate the humoral immune system. Consistently aberrant BAFF-R expression in leukemic cells reveals an intimate connection of these cells’ malignant physiology to the BAFF/BAFF-R axis and also provides an additional survival mechanism to the expressing cells. In this study, we used primary cells and cell lines to interrogate the mechanisms underlying aberrant BAFF-R expression in precursor B acute lymphoblastic leukemia (precursor B-ALL) and mature B chronic lymphocytic leukemia (CLL). Here we demonstrate the aberrant expression of BAFF-R in precursor B-ALL cell lines and reveal that these cells acquire BAFF-R expression through premature transcriptional activation of the BAFF-R promoter in coordination with regulatory transcription factor c-Rel. Investigations using primary CLL cells provide a crucial counterpoint through their paucity of BAFF-R relative to their benign mature B cell counterparts, which we establish as functionally significant in its depletion of the CLL cells’ BAFF-binding capacity. Furthermore, BAFF-R downregulation in CLL patients is revealed here to be restricted to the malignant compartment and mediated post-transcriptionally in order to compensate for the consistently unchanged levels of transcription factor c-Rel and BAFF-R mRNA. Finally, we present evidence that CLL cells retain endogenous mechanisms of BAFF-R regulatory control despite active receptor dysregulation.
Introduction
The implications of dysregulation of a receptor capable of imparting survival and costimulatory signals are vast. The ability to manipulate such a powerful axis may impart, or at least signify, independence of environmentally-regulated homeostatic control. This phenomenon has been largely overlooked in the regulation of survival and proliferation-enhancing TNF family receptor BAFF-R, the receptor component of the BAFF/BAFF-R cytokine/receptor axis.
Investigations of the relationship between BAFF and B cell-driven pathologies have primarily focused on the levels of BAFF provided to the pathological cells either by the immune environment or an autocrine mechanism. Several studies have demonstrated a correlation between BAFF production and autoimmune pathology in diseases like systemic lupus erythematosus (SLE) and rheumatoid arthritisCitation1–Citation6 and have demonstrated a positive effect of BAFF on pathological autoimmune and malignant cells in vitro.Citation7–Citation9 In neoplastic diseases like chronic lymphocytic leukemia (CLL), characterized by a proliferative, clonal, aberrant B cell, however, it is less clear to what extent BAFF contributes as a cause of more serious disease. While evidence linking higher circulating BAFF levels to familial CLL raises the possibility of a causative link,Citation10 several reports have demonstrated a drop in the circulating BAFF levels of sporadic CLL patients relative to normal controls.Citation11–Citation14 Although a decrease in CLL patients' circulating BAFF levels damages the hypothesis that BAFF plays a role in tumorigenesis, it is consistent with the malignant clone's reduction of soluble BAFF levels through its active use of BAFF-R. While most evidence supports the active exploitation of a BAFF-mediated survival pathway in CLL and other BAFF-R-expressing B lineage malignancies, expression of the receptor may significantly dampen normal humoral immunity by acting as a sink for this crucial survival molecule even if the malignancies are entirely divorced from normal BAFF physiology. Likewise in precursor B lymphoblastic leukemias/lymphomas (precursor B-ALLs) that aberrantly express BAFF-R, the receptor may coopt normal BAFF physiologic mechanisms to grant malignant cells an additional survival pathway and also hinder normal humoral immunity through competition for BAFF.
In addition to observed associations of BAFF with CLL and CLL survival,Citation9,Citation10,Citation13–Citation15 several studies have demonstrated the expression of BAFF-R by CLL cells.Citation9,Citation13,Citation16 While Briones et al. previously recognized a decrease in biotinylated BAFF binding to CLL cells compared to their normal counterparts,Citation17 a decrease in surface BAFF-R levels in CLL compared to normal cells was not reported until recently. Lin et al. reported data showing a trend of lower BAFF-R expression in CLL cells, but neglected to describe the trend's potential significance or even to note its existence in the text.Citation18 In this study, we extend these findings to generalize the phenomenon of BAFF-R downregulation to the vast majority of CLL cases and for the first time, determine the point at which regulation occurs.
The expression of BAFF-R by cancers that arise from tissues that do not normally express it may both directly aid malignant cell survival and dampen normal humoral immunity through BAFF-binding. Our studies reported here using precursor B-ALL-derived cell lines support the recently published finding that precursor B cell cancers aberrantly express BAFF-R.Citation19 These cell lines enabled us to study the mechanisms of expression in a straight-forward way with abundant, manipulatable material.
The studies described here demonstrate the dysregulation of BAFF-R in B cell cancers: a downregulation of surface BAFF-R in CLL and the aberrant expression of surface BAFF-R in precursor B-ALL. These findings are supported by an analysis of BAFF-R-regulatory molecule c-Rel expression in these same cells and together reveal complexities in the altered relationships between these malignant cells and the immune system relevant to the treatment of both malignancies and malignancy-associated humoral immunodeficiencies.
Results
Malignant early B cells aberrantly express BAFF-R.
While the BAFF/BAFF-R cytokine axis has gained significant prominence as a survival mechanism both for normal BCR-expressing B cells and for their malignant counterparts, it is only recently that a report has identified a place for the axis in precursor B cell cancers.Citation19 This report encouraged us to pursue the observation that the early B cell lines Reh and NALM-6, derived from patients with precursor B-ALL and proposed to represent malignant counterparts to pro-B and pre-B cells, respectively, express surface BAFF-R ().
Based on previous work demonstrating a link between NFκB family member c-Rel and BAFF-R expression, we used qRT-PCR to ascertain the mRNA levels of c-Rel (). The standing levels of mRNA for c-Rel were similar in three of the tested cell lines and significantly decreased in the least mature line, Reh. While this decrease in message levels is consistent with Reh's distinction as the least BAFF-R surface expressing line, it was unexpected that it would differ to such an extent from NALM-6, which shares a similar level of BAFF-R surface expression with Reh but shares a c-Rel mRNA signature with the mature B cell lines.
The discrepancy between c-Rel levels in the two precursor B cell lines led us to pursue another layer of control in the c-Rel transcriptional regulatory axis: nuclear localization. Immunoblotting for c-Rel levels in the nuclear fraction of protein extracts from the four cell lines () revealed that while the mRNA levels of c-Rel differed between the two precursor lines, the amount of nuclear c-Rel protein did not and is equally depressed relative to the nuclear c-Rel in the two mature B cell lines.
Malignant precursor B cell lines activate the TNFRSF13C promoter.
The discovery that precursor B cell lines Reh and NALM-6 express BAFF-R and may regulate c-Rel to do so led us to extend our previous work identifying the BAFF-R gene TNFRSF13C's promoter to the precursor B cell lines. Our previous work demonstrated that the 0.5 kb genomic sequence upstream of TNFRSF13C was sufficient to account for the bulk of promoter activity in BAFF-R-expressing mature B cell lines and the absence of activity in plasma cell lines that do not express BAFF-R.Citation20 By transfecting the 0.5 kb TNFRSF13C promoter luciferase reporter into both the precursor B cell lines and the mature B cell lines, we were able to determine that there was not a significant difference in the extent to which the precursor lines and the mature B cell lines could activate the promoter (). While the extent to which the various lines used the promoter to increase firefly luciferase expression and the statistical significances thereof differed between cell lines, in both precursor B cell lines the increase in expression of the promoter vector over empty vector was at least 2.8-fold in each individual transfection.
CLL downregulation of surface BAFF-R functionally impairs BAFF binding.
The aberrant expression of BAFF-R, as in the malignant precursor B cell lines Reh and NALM-6 (), is not the rule among B lineage derived cancers. In fact, we have observed the opposite, significant downregulation of BAFF-R, on the surface of primary CLL populations (). CLL cells, which arise from a normally BAFF-R-expressing mature B cell population, are shown divided into two categories, mutated and unmutated. These categories stratify patients into two levels of risk of progression and refer to whether the immunoglobulin heavy chain variable region gene shows evidence of somatic hypermutation. CLL populations with an unmutated phenotype are at a higher risk of progression than those with a mutated phenotype.Citation21,Citation22 In this case, the mutation phenotype did not reveal differences in BAFF-R expression.
To determine the extent to which the decrease in surface BAFF-R impaired BAFF binding, we stained the cells for bound BAFF immediately following isolation from blood and also after incubation with an excess of recombinant BAFF (). Under these circumstances, it was clear that not only did normal B cell populations have much more BAFF bound initially, but they also had a much greater BAFF-binding capacity than the CLL populations. Although the CLL populations had uniformly lower BAFF-R levels and lower levels of endogenous bound BAFF, many also showed a limited amount of additional BAFF binding following incubation with recombinant BAFF, indicating the presence of residual BAFF-binding capacity, as was present to a much greater degree in all the normal B cell populations ().
Downregulation of BAFF-R in CLL is restricted to the malignant clone.
Despite the consistent observation that CLL B cells show low BAFF-R surface expression and low BAFF binding, there has been little analysis as to whether the decrease is unique to the B cells of the CLL clone or to all of the CLL patient's B cells.Citation18 It is likely that this has been overlooked in part due to the difficulty inherent in examining the normal B cell population in CLL, which is dwarfed by the malignant clone even at the time the cancer is initially diagnosed. To determine whether the aberrant BAFF-R surface expression represented a defect of the CLL clone or of the CLL patient's humoral immune system more generally, we used flow cytometry to identify the normal (CD19+/CD5−) and malignant (CD19+/CD5+) B cell populations within CLL patients' peripheral blood and co-stained for BAFF-R or an isotype control (). In each case, the BAFF-R level was greater on the normal population than on the malignant clone and showed levels comparable to normal peripheral blood B cells ().
Decreases in CLL BAFF-R surface expression are post-transcriptional.
After establishing the BAFF-binding impairment coincident to lower BAFF-R surface expression, we wished to establish the source of the receptor dysregulation. While the pre-B and mature B cell lines demonstrated an association between nuclear c-Rel levels and surface BAFF-R expression consistent with established TNFRSF13C promoter regulation (), the CLL cells showed a surprising trend: the BAFF-Rlow CLL populations had c-Rel and BAFF-R mRNA levels equal to or greater than those of the BAFF-Rnormal/hi normal control B cell populations (). While the mRNA levels could not explain the difference in BAFF-R surface expression (), the c-Rel and BAFF-R mRNA levels () still demonstrated a relationship consistent with c-Rel-mediated TNFRSF13C promoter regulation established previously in reference Citation20. Applying linear regression analysis to the c-Rel and BAFF-R mRNA levels in all samples or either of the CLL subpopulations showed a significant correlation between the expression of these two genes (all samples: r2 = 0.91, p < 0.0001; mutated: r2 = 0.99, p < 0.0001; unmutated r2 = 0.97, p < 0.0001).
Despite the clear drop in BAFF-R surface expression that initiated this line of inquiry, the mRNA levels in CLL and normal peripheral blood B cells did not significantly differ, but instead trended toward normal or higher BAFF-R mRNA content in CLL samples compared to normal B cells. This seeming inconsistency required us to examine the absolute protein levels of BAFF-R in the cells. Immunoblotting with five representative CLL samples and four normal B cell samples () revealed that the total protein levels were consistent with the observed surface expression in these categories of cells. The levels of BAFF-R protein identified by immunoblot were similar to one another and mostly lower in the CLL samples, but there was some overlap with the least BAFF-R-expressing of the four normal B cell samples. The three other normal samples showed similar and significantly higher levels of BAFF-R protein expression. The decrease in total BAFF-R protein despite normal to increased mRNA levels confirmed the regulation to be post-transcriptional.
CLL cells actively regulate BAFF-R expression.
The consistently lower levels of BAFF-R surface expression in malignant CLL B cells suggest that the cells have become independent of BAFF-R and the normal mechanisms controlling BAFF-R expression have been lost to the clone during its genetic evolution. In order to determine whether the CLL clone has retained the capacity to regulate BAFF-R expression, we stimulated CLL cells with CpG, a polyclonal stimulus shown to induce BAFF-R downregulation in normal B cell populations. While the CLL cells did not uniformly respond to the CpG stimulation, all of them showed large populations with decreasing surface BAFF-R, and all but one showed a very tight histogram with a single peak (). In the differing case, the CLL cells showed a range of BAFF-R expression from a lower level up to the level at which the unstimulated cells express it, reflecting a diversity of response within the CLL population.
Discussion
The BAFF/BAFF-R survival axis has been explored largely in two contexts in B lineage cancers: (1) defining the ability of the malignant cells to exploit this endogenous survival pathway and (2) identifying chemotherapeutic agents that directly target the BAFF ligand or the BAFF receptor. In this study, we take a third approach that identifies a crucial dimension of the malignant manipulation of the BAFF/BAFF-R survival axis by exploring the dysregulation of BAFF-R in malignant B cell populations. Understanding the way that B lineage leukemias access and exploit this cytokine survival axis through receptor regulation provides significant insight into the physiology of these cancers and modes of therapeutic intervention.
Our data demonstrating the expression of BAFF-R on the surface of precursor B-ALL-derived precursor B cell lines Reh and NALM-6 () are consistent with a recent report demonstrating the presence of BAFF-R on the surface of primary precursor B-ALL cells.Citation19 We were encouraged to pursue this observation as reflective of a striking and unexpected phenomenon common to B lineage ALL malignancies. This aberrant BAFF-R expression may indeed fuel the growth of these cancers and even play a role in their tumorigenesis, as their normal counterparts entirely lack expression of this anti-apoptotic and costimulatory cytokine receptor.
To determine how these cells had upregulated BAFF-R, we examined the levels of one crucial regulatory transcription factor, c-Rel, established to play a role in the promotion of BAFF-R expression.Citation20 Immunoblotting of nuclear c-Rel showed similar levels in the two precursor lines, distinctly lower than the similar and higher levels of the two mature lines. These nuclear c-Rel levels support the relevance of c-Rel as a regulatory transcription factor for BAFF-R expression, as the nuclear protein levels are concordant with the levels of surface expression of BAFF-R in these four lines, i.e., the two precursor lines show lower c-Rel nuclear protein and lower surface BAFF-R, while the two mature lines show higher nuclear c-Rel protein and higher BAFF-R surface expression.
Employing a reporter system with the 0.5 kb upstream of the TNFRSF13C gene, we then assessed the ability of this region to promote firefly luciferase expression in the pre-B lines, as it can in mature B cell lines Loukes and RAMOS. The differences between the resulting light production in the four lines were not statistically significant, suggesting that these two precursor B cell lines are just as capable of promoting the BAFF-R gene as the mature lines, though the extent of this ability varies. The implication of these results is that these two early B cell lines have acquired the autonomous ability to promote BAFF-R expression. Whereas normal developing B cells acquire surface BAFF-R expression only after the emergence of a complete, functional B cell receptor on the cell surface, these precursors, which lack surface immunoglobulin, promote the BAFF-R gene nonetheless. The mechanism fueling aberrant BAFF-R expression in precursor B-ALL cancers may provide significant insight into the common physiology underlying the disease. The observation of a correlation with nuclear c-Rel expression is consistent with a mechanism of aberrant transcription factor activation, but the complexity of the transcription factor network in these cells and the possibility of epigenetic regulation necessitate significant further investigation. While primary precursor B-ALL cells have been shown to be capable of exploiting soluble BAFF for survival enhancement in vitro,Citation19 the independence of these cell lines from exogenous BAFF brings the necessity of the receptor into question. However, the possibilities remain that an undetected autocrine BAFF loop is enabling survival, or that a previously active and essential BAFF/BAFF-R axis allowed these cells to escape normal homeostasis during tumorigenesis.
BAFF-R downregulation in CLL is a valuable counterpoint to the BAFF-R upregulation characteristic of precursor B-ALL. In CLL, not only are levels of surface BAFF-R distinctly lower than normal B cells on average, but in all examined cases, the CLL cells had less surface-bound BAFF and less BAFF-binding capacity than their normal counterparts. The downregulation of BAFF-R and the accompanying loss of BAFF binding suggest that these cells have gained an independence of this cytokine axis and subsequently lost BAFF-R expression stochastically during the process of clonal evolution, but the universality and uniformity of the receptor decrease in these cells are the hallmarks of a more specific, regulated process. The loss of expression may represent changes in processing secondary to the induction of an autocrine pathway or an as yet unrecognized BAFF-Rlow cell of origin, but the regulated nature of the decrease and its consistency across patients distinctly suggest that the mechanism of BAFF-R downregulation shares a single, consistent etiology.
Perhaps the simplest explanation for the BAFF-R downregulation would be the external, global pressure placed on the BAFF-R expressing cells by the alterations in BAFF-mediated homeostasis imposed by a tremendous abnormal B cell population. In the case that the CLL BAFF-R downregulation represents a normal response to these abnormal conditions, the normal B cell compartment should reflect this shift in BAFF-R expression. However, the categorically higher level of BAFF-R surface levels in these accompanying small normal B cell populations suggest that this is not a physiologic response to changes in homeostatic control but rather a quality specific to the malignant clone.
Regardless of the provenance of the low BAFF-R surface expression among CLL cells, the mechanisms of the change in expression offer insight into the physiology of the malignant cells. Unlike the direct relationship between nuclear c-Rel, BAFF-R mRNA and BAFF-R surface expression in B cell development and in precursor B-ALL, BAFF-R downregulation does not stem from a transcriptional decrease in BAFF-R in CLL. qRT-PCR analysis showed that levels of both BAFF-R and a defining transcription factor, c-Rel, were normal to elevated in the CLL cells relative to normal mature B cells. While the nearly linear relationship between c-Rel and BAFF-R mRNA levels supported the regulatory nature of c-Rel in BAFF-R transcription, the normal to increased levels of BAFF-R mRNA in CLL over normal B cells ran counter to the decrease in BAFF-R surface expression. Immunoblotting for BAFF-R revealed that the total protein levels were indeed decreased in the CLL cells compared to normal B cells. The low total levels of protein in CLL despite the normal to increased mRNA show that post-transcriptional regulation tempers the translation of BAFF-R mRNA and keeps absolute BAFF-R protein levels categorically depressed in the malignant cells. Whether this post-transcriptional regulatory mechanism acts directly on translation, as a specific microRNA (miR) would, or reflects the rapid degradation of translated or even post-signaling protein remains to be seen. The identification of regulation as occurring at the post-transcriptional level and as unique to the CLL clone suggests that downregulation is an abnormality intrinsic to the malignant clone.
Despite the CLL cells' intrinsic BAFF-R downregulatory ability, CpG activation stimulated the malignant cells and induced further BAFF-R downregulation, just as it does in normal B cells. The retention of this endogenous mechanism of receptor control signifies the maintenance of physiologic pathways and reveals another layer of similarity of the malignant clone to the normal B cell compartment. Defining the relationships between the malignant clone and the normal compartment is crucial in directing therapy to the cancer: while targeting a shared pathway may indeed be useful, targeting only the process unique to the cancer, in this case targeting a malignancy-derived post-transcriptional mechanism, may both harm the cancer and spare the healthy humoral immune system.
In summary, these studies reveal the regulatory levels at which two B lineage cancers dysregulate BAFF-R surface expression. In the aberrantly BAFF-R-expressing precursor B cell malignancies of precursor B-ALL, nuclear c-Rel correlates with BAFF-R expression levels, while in the aberrantly BAFF-Rlow mature B cell malignancy CLL, the drop in surface BAFF-R is post-transcriptional. In both cases, a clearer understanding of control provides insight into the pathophysiology of these malignancies and identifies novel targets for therapeutics in c-Rel and post-transcriptional mediators.
Materials and Methods
Ethics statement.
Blood was collected from CLL patients after informed consent and Mayo Clinic Institutional Review Board approval and in accord with the Declaration of Helsinki. Informed consent was not required for the healthy donor material as this material is considered by the Institutional Review Board as waste material generated during blood donation. In addition, healthy donor samples arrive de-identified in the laboratory.
Cells and cell lines.
Peripheral blood mononuclear cells (PBMCs) from CLL patients were separated from heparinized venous blood by Ficoll-Hypaque density gradient centrifugation. The CLL PBMCs used in this study were generally greater than 90% CLL B cells (CD19+/CD5+). Normal peripheral blood B cells were isolated from plasma apheresis cones by Ficoll-Hypaque density gradient centrifugation followed by purification with a negative selection protocol (StemCell Technologies) using a RoboSep. Normal B cells were routinely greater than 98% pure as revealed by CD19 expression. Leukemic cells were cultured in serum-free adoptive immunotherapy media-V (AIM-V, Gibco-Invitrogen). In all cases, fresh samples were used. For activation experiments, cells were cultured for 48 hours in media containing 2.5 µg/ml CpG ODN 200619 (synthesized in house), 9.4 × 104 IU/ml human IL-2 and 10 ng/ml human IL-15 (both Peprotech).
EBV-negative Burkitt's lymphoma B cell lines RAMOS and Loukes and precursor B-ALL-derived precursor B cell lines Reh and NALM-6 were maintained in RPMI-1640 supplemented with 10% heat-inactivated FCS, penicillin, streptomycin, glutamine and gentamicin.
Flow cytometric analyses.
Cell lines, normal peripheral blood B cells, and CLL cells were routinely stained for BAFF-R using a BAFF-R-PE antibody and comparable isotype control antibody (both eBioscience) in all experiments. CD19-APC, CD5-PE (both BD Biosciences), and BAFF-R-FITC or IgG2a/κ-FITC isotype control antibodies (both eBioscience) were used to analyze CLL samples for BAFF-R expression on the malignant and normal B cell subpopulations. The antibodies were incubated with the cells for 25 min at 4°C before two washes and fixation in PBS containing 1% paraformaldehyde. Data were collected on a FACS Vantage or Aria (BD Biosciences) and analyzed with Flow Jo software (Tree Star).
In the BAFF binding assays, cells were stained immediately for BAFF and BAFF-R or incubated with recombinant BAFF (Alexis, Enzo Life Sciences, Inc.,) at 500 ng/mL on ice for 20 minutes before BAFF staining. Cells were stained with a biotinylated anti-BAFF Ab or a biotinylated control antibody (both R&D Systems) 25 minutes on ice, washed twice, incubated with a streptavidin-PE secondary antibody (Caltag-Invitrogen), washed twice and fixed with 1% paraformaldehyde.
Real-time quantitative RT-PCR for C-Rel and BAFF-R.
RNA was isolated from cells using TRIzol reagent (Invitrogen) followed by phenol:chloroform extraction, isopropanol precipitation and a 70% ethanol wash. 2 µg of RNA were converted to cDNA with the 1st Strand cDNA Synthesis Kit (GE Healthcare). The cDNA was diluted to 50 µL with diethylpyrocarbonate-treated molecular grade water before using 2 µL in each real-time quantitative RT-PCR (qRT-PCR) reaction performed in duplicate with RT2 SYBR green/Rox qPCR Master Mix (SABiosciences) in an ABI Prism 7900HT Real Time System. cDNA was diluted 1:200 before use in the 18S control reaction to preserve the linearity of amplification in an appropriate threshold cycle range greater than 10. C-Rel was amplified with primers forward 5′-CCT GTT GTC TCG AAC CCA AT and reverse 5′-TCT CCT CCT CTG ACA CTT CC. BAFF-R was amplified with primers forward 5′-GGT CCT GGT GGG TCT GGT GAG and reverse 5′-ACC TTG TCC AGG GGC TCT GGG. 18S rRNA was amplified with primers forward 5′-CGG CTA CCA CAT CCA AGG AA and reverse 5′-GCT GGA ATT ACC GCG GCT.
Immunoblotting.
Nuclear extracts were prepared with NE-PER Nuclear and Cytoplasmic Extraction reagents (Pierce) and whole cell lysates were prepared with RIPA buffer. Protein was resolved by SDS-PAGE (50 µg/lane for c-Rel and 20 µg/ lane for BAFF-R) and transferred to Immobilon-P membranes (Millipore) for immunoblotting. Membranes were blocked for 1 h at 21°C in 5% Blotto (Santa Cruz Biotechnology) supplemented with 0.2% Tween 20 and then blotted overnight with anti-c-Rel Ab (Millipore), anti-Histone H1 Ab (Millipore), anti-BAFF-R Ab (ProSci), or anti-β-Actin Ab (Novus Biologicals) following the manufacturer's protocol. Immunoreactive proteins were detected using an ECL detection system (Super Signal; Pierce) and autoradiography.
Luciferase reporter constructs and transient transfections.
Reporters were generated and transfections were carried out as previously published in reference Citation20.
Statistical analysis.
Within the luciferase reporter assays, ANOVA revealed no significant difference between the activities of the TNFRSF13C promoter in the four different cell lines, while two-tailed student's t-tests were used to compare the activity of the promoter to the empty reporter. Means in through were compared with ANOVA and two-tailed student's t-tests where appropriate. Linear regression was performed using all the data points presented in as well as mutated and unmutated subsets. In all cases, p < 0.05 was considered significant unless otherwise specified.
Abbreviations
| Ab | = | antibody |
| AIM-V | = | adoptive immunotherapy media-V |
| ANOVA | = | analysis of variance |
| APC | = | allophycocyanin |
| BAFF | = | B cell activating factor of the TNF family |
| BAFF-R | = | BAFF-receptor |
| BCR | = | B cell receptor |
| cDNA | = | complementary DNA |
| CLL | = | chronic lymphocytic leukemia |
| EBV | = | epstein-barr virus |
| ECL | = | enhanced chemiluminescence |
| FACS | = | fluorescence-activated cell sorting |
| FCS | = | fetal calf serum |
| FITC | = | fluorescein isothiocyanate |
| MFI | = | mean fluorescence intensity |
| ODN | = | oligodeoxynucleotide |
| PBMC | = | peripheral blood mononuclear cell |
| PCR | = | polymerase chain reaction |
| PE | = | phycoerythrin |
| precursor B-ALL | = | precursor B acute lymphoblastic leukemia |
| qPCR | = | quantitative PCR |
| qRT-PCR | = | real-time quantitative RT-PCR |
| RIPA | = | radio-immunoprecipitation assay |
| RT-PCR | = | reverse transcription PCR |
| SDS-PAGE | = | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SLE | = | systemic lupus erythematosus |
| TNF | = | tumor necrosis factor |
Figures and Tables
Figure 1 B cell lines derived from early B cell cancers aberrantly express BAFF-R. (A) Flow cytometric analysis of BAFF-R staining (empty histogram) and isotype control staining (filled histogram) in precursor B-ALL-derived cell lines Reh and NALM-6 compared to the Burkitt's lymphoma-derived mature B cell lines Loukes and RAMOS. Ratio of geometric mean fluorescence intensity of specific staining to isotype control staining (ΔMFI) is inset. (B) Quantitative RT-PCR analysis of c-Rel mRNA levels in the precursor B-ALL and Burkitt's lymphoma lines. (C) Immunoblot for c-Rel and nuclear loading control Histone H1. Approximately 50 µg of nuclear extracts were loaded and blotted for the labeled proteins.
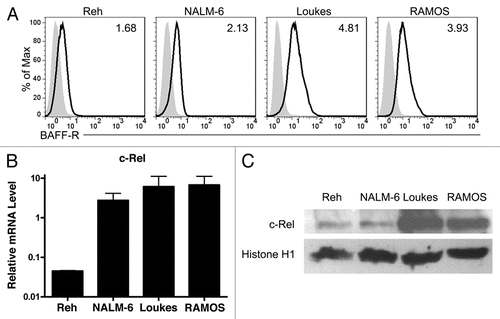
Figure 2 BAFF-R-expressing early B cell lines show significant BAFF-R promoter reporter activity. The genomic region spanning the 0.5 kb upstream of TNFRSF13C, the gene encoding BAFF-R, ending 6 bp 5′ of the start codon, was cloned and inserted into firefly luciferase reporter vector pGL3-Basic. This reporter was co-transfected into the target cell lines by electroporation with a fixed amount of control Renilla luciferase reporter vector pRL-TK. Specific promoter activity is reported as relative light units, the ratio of firefly: Renilla luciferase activity normalized to the relative activity of the empty pGL3-Basic vector. **p < 0.01.
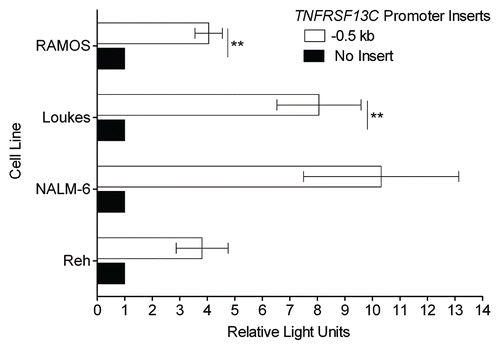
Figure 3 CLL cells lose significant BAFF-binding capacity coincident with downregulation of BAFF-R surface expression. (A) Normal B cells and CLL cells were stained for BAFF-R and an isotype-matched control antibody. The ratio of the geometric mean fluorescence intensity in specific to control antibody staining is reported here (ΔMFI). CLL samples were divided into mutated and unmutated groups reflecting the amount of somatic hypermutation undergone at the immunoglobulin locus as determined by sequencing. (B) The same CLL samples tested for BAFF-R expression in (A) and independent normal B cell controls were tested for BAFF binding by incubation with an anti-BAFF antibody labeled with biotin and stained secondarily with streptavidin bound to fluorophore. ΔMFI was determined as above relative to an isotype-matched control primary antibody. BAFF-binding was determined both immediately after isolation (“Endogenous BAFF”) and after 20 minute incubation with an excess of recombinant BAFF (“Exogenous BAFF”). (C) BAFF-binding in the normal and CLL B cells represented as matched pairs before and after the addition of recombinant BAFF. *p < 0.05; **p < 0.01; ***p < 0.001.
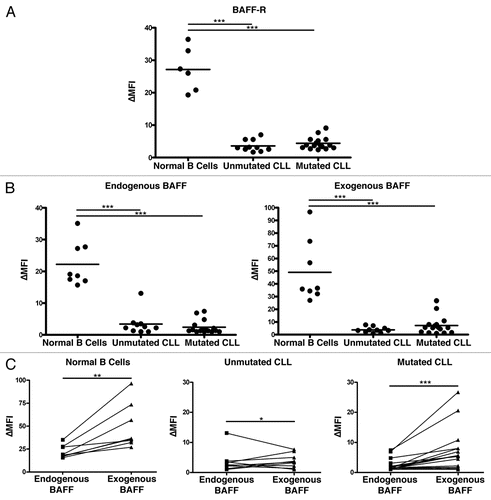
Figure 4 The normal CD5− B cell compartment in CLL patients retains greater BAFF-R expression than the malignant clone. (A) Representative histogram of normal peripheral blood B cells (far right histogram) and malignant CLL cells (center and left histograms) isolated with Ficoll-Hypaque separation and co-stained with antibodies to CD19, CD5 and either BAFF-R (black outlined histograms) or an isotype-matched control antibody (filled histogram without outline). (B) Matched CD5+ (CLL) and CD5− (normal peripheral blood B) cell BAFF-R expression relative to the isotype-matched control antibody, ΔMFI. *p < 0.05.
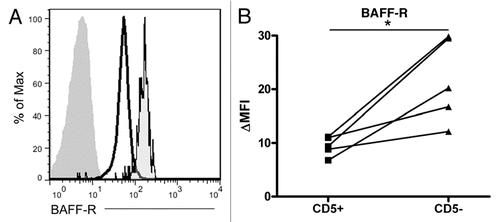
Figure 5 The regulation of BAFF-R expression in CLL is primarily post-transcriptional. (A) Relative expression of BAFF-R and NFκB family member c-Rel in CLL and normal peripheral blood B cells. Expression levels were determined by qRT-PCR and normalized to 18S rRNA. (B) The relationship between c-Rel and BAFF-R mRNA levels in CLL and normal B cells. (C) Immunoblot for BAFF-R and β-actin in five CLL and four normal peripheral blood B cell samples. Densitometric analysis was performed and quantified as a ratio of adjusted intensity volume of BAFF-R to β-actin and normalized to a scale of 0 to 10. Relative values are noted above the BAFF-R blot.
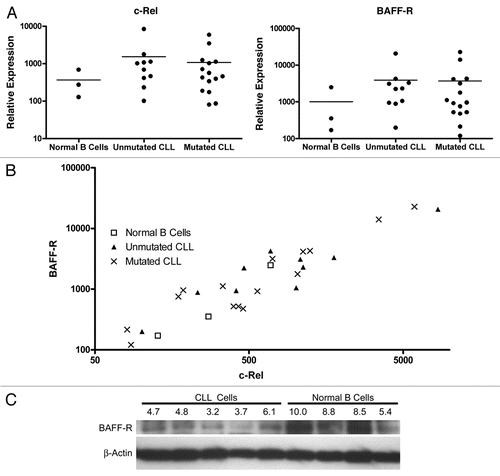
Figure 6 CLL cells employ native BAFF-R regulatory pathways in response to CpG stimulation. Isolated CLL cells were stimulated with CpG and IL-2 in vitro for two days (empty histogram) or not stimulated (dark gray histogram) and stained for BAFF-R expression or an isotype-matched control antibody (light gray histogram).
Acknowledgements
We thank Dr. Neil Kay for his illuminating discussion. This work was supported by the National Institutes of Health CA105258 and CA062242 (awarded to D.F.J.).
References
- Cheema GS, Roschke V, Hilbert DM, Stohl W. Elevated serum B lymphocyte stimulator levels in patients with systemic immune-based rheumatic diseases. Arthritis Rheum 2001; 44:1313 - 1319
- Collins CE, Gavin AL, Migone TS, Hilbert DM, Nemazee D, Stohl W. B lymphocyte stimulator (BLyS) isoforms in systemic lupus erythematosus: disease activity correlates better with blood leukocyte BLyS mRNA levels than with plasma BLyS protein levels. Arthritis Res Ther 2006; 8:6
- Mariette X, Roux S, Zhang J, Bengoufa D, Lavie F, Zhou T, et al. The level of BLyS (BAFF) correlates with the titre of autoantibodies in human Sjogren's syndrome. Ann Rheum Dis 2003; 62:168 - 171
- Zhang J, Roschke V, Baker KP, Wang Z, Alarcon GS, Fessler BJ, et al. Cutting edge: a role for B lymphocyte stimulator in systemic lupus erythematosus. J Immunol 2001; 166:6 - 10
- Tan SM, Xu D, Roschke V, Perry JW, Arkfeld DG, Ehresmann GR, et al. Local production of B lymphocyte stimulator protein and APRIL in arthritic joints of patients with inflammatory arthritis. Arthritis Rheum 2003; 48:982 - 992
- Chu VT, Enghard P, Schurer S, Steinhauser G, Rudolph B, Riemekasten G, et al. Systemic activation of the immune system induces aberrant BAFF and APRIL expression in B cells in patients with systemic lupus erythematosus. Arthritis Rheum 2009; 60:2083 - 2093
- Petri M, Stohl W, Chatham W, McCune WJ, Chevrier M, Ryel J, et al. Association of plasma B lymphocyte stimulator levels and disease activity in systemic lupus erythematosus. Arthritis Rheum 2008; 58:2453 - 2459
- Doreau A, Belot A, Bastid J, Riche B, Trescol-Biemont MC, Ranchin B, et al. Interleukin 17 acts in synergy with B cell-activating factor to influence B cell biology and the pathophysiology of systemic lupus erythematosus. Nat Immunol 2009; 10:778 - 785
- Endo T, Nishio M, Enzler T, Cottam HB, Fukuda T, James DF, et al. BAFF and APRIL support chronic lymphocytic leukemia B-cell survival through activation of the canonical NFkappaB pathway. Blood 2007; 109:703 - 710
- Novak AJ, Grote DM, Ziesmer SC, Kline MP, Manske MK, Slager S, et al. Elevated serum B-lymphocyte stimulator levels in patients with familial lymphoproliferative disorders. J Clin Oncol 2006; 24:983 - 987
- Planelles L, Castillo-Gutierrez S, Medema JP, Morales-Luque A, Merle-Beral H, Hahne M. APRIL but not BLyS serum levels are increased in chronic lymphocytic leukemia: prognostic relevance of APRIL for survival. Haematologica 2007; 92:1284 - 1285
- Haiat S, Billard C, Quiney C, Ajchenbaum-Cymbalista F, Kolb JP. Role of BAFF and APRIL in human B-cell chronic lymphocytic leukaemia. Immunology 2006; 118:281 - 292
- Kern C, Cornuel JF, Billard C, Tang R, Rouillard D, Stenou V, et al. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood 2004; 103:679 - 688
- Bojarska-Junak A, Hus I, Chocholska S, Wasik-Szczepanek E, Sieklucka M, Dmoszynska A, et al. BAFF and APRIL expression in B-cell chronic lymphocytic leukemia: correlation with biological and clinical features. Leuk Res 2009; 33:1319 - 1327
- Molica S, Digiesi G, Mauro F, Mirabelli R, Cutrona G, Vitelli G, et al. Increased serum BAFF (B-cell activating factor of the TNF family) level is a peculiar feature associated with familial chronic lymphocytic leukemia. Leuk Res 2009; 33:162 - 165
- Novak AJ, Bram RJ, Kay NE, Jelinek DF. Aberrant expression of B-lymphocyte stimulator by B chronic lymphocytic leukemia cells: a mechanism for survival. Blood 2002; 100:2973 - 2979
- Briones J, Timmerman JM, Hilbert DM, Levy R. BLyS and BLyS receptor expression in non-Hodgkin's lymphoma. Exp Hematol 2002; 30:135 - 141
- Lin WY, Gong Q, Seshasayee D, Lin Z, Ou Q, Ye S, et al. Anti-BR3 antibodies: a new class of B-cell immunotherapy combining cellular depletion and survival blockade. Blood 2007; 110:3959 - 3967
- Parameswaran R, Muschen M, Kim YM, Groffen J, Heisterkamp N. A functional receptor for B-cell-activating factor is expressed on human acute lymphoblastic leukemias. Cancer Res 2010; 70:4346 - 4356
- Mihalcik SA, Huddleston PM 3rd, Wu X, Jelinek DF. The Structure of the TNFRSF13C Promoter Enables Differential Expression of BAFF-R during B Cell Ontogeny and Terminal Differentiation. J Immunol 2010; 185:1045 - 1054
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94:1840 - 1847
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94:1848 - 1854