Abstract
The Ins(1,4,5)P3 receptor acts as a central hub for Ca2+ signaling by integrating multiple signaling modalities into Ca2+ release from intracellular stores downstream of G-protein and tyrosine kinase-coupled receptor stimulation. As such, the Ins(1,4,5)P3 receptor plays fundamental roles in cellular physiology. The regulation of the Ins(1,4,5)P3 receptor is complex and involves protein-protein interactions, post-translational modifications, allosteric modulation, and regulation of its sub-cellular distribution. Phosphorylation has been implicated in the sensitization of Ins(1,4,5)P3-dependent Ca2+ release observed during oocyte maturation. Here we investigate the role of phosphorylation at T-930, a residue phosphorylated specifically during meiosis. We show that a phosphomimetic mutation at T-930 of the rat Ins(1,4,5)P3 receptor results in decreased Ins(1,4,5)P3-dependent Ca2+ release and lowers the Ins(1,4,5)P3 binding affinity of the receptor. These data, coupled to the sensitization of Ins(1,4,5)P3-dependent Ca2+ release during meiosis, argue that phosphorylation within the coupling domain of the Ins(1,4,5)P3 receptor acts in a combinatorial fashion to regulate Ins(1,4,5)P3 receptor function.
Introduction
Ca2+ release from intracellular stores is mediated by two major classes of intracellular channels, ryanodine and Ins(1,4,5)P3 receptors. Mammalian genomes encode three Ins(1,4,5)P3 receptor genes, type 1–3, with disparate tissue distribution and molecular regulation.Citation1 The Ins(1,4,5)P3 receptor is a complex signal integrator that consolidates input from multiple signaling cascades into Ca2+ release from intracellular stores.Citation2 The Ins(1,4,5)P3 receptor (~2700 amino acids) is functionally divided into three domains: the Ins(1,4,5)P3 binding domain, the coupling domain and the channel domain (). The pore forming domain localizes to the C-terminal end of the protein and is composed of six membrane spanning regions and a pore forming loop between trans-membrane domains 5 and 6, followed by a short cytoplasmic tail. The Ins(1,4,5)P3 binding domain in the N-terminus of the protein binds the ligand Ins(1,4,5)P3, and the coupling region spans the region between the Ins(1,4,5)P3 binding region and the channel domain.Citation2,Citation3 The coupling domain transmits Ins(1,4,5)P3 binding into gating of the channel to release Ca2+ from the endoplasmic reticulum (ER). In addition to Ins(1,4,5)P3, Ca2+ functions as a co-agonist of the Ins(1,4,5)P3 receptor regulating it’s gating in a biphasic fashion.Citation4,Citation5 The bulk of the Ins(1,4,5)P3 receptor protein is cytoplasmic and offers multiple sites for regulation by interacting proteins, small molecules and post-translational modifications.Citation1,Citation2
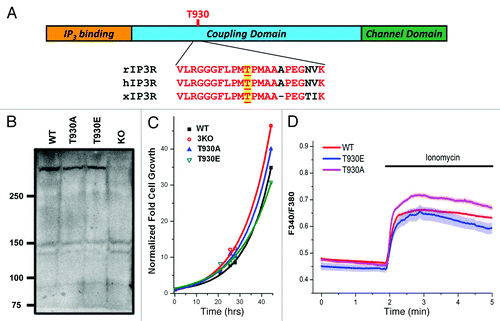
Figure 1. Generation and characterization of stable DT40 cell lines expressing Ins(1,4,5)P3 receptor phosphorylation mutants. (A) Cartoon representation of Ins(1,4,5)P3 receptor structure with sequence alignment around T-930, for the rat (rIP3R), human (hIP3R) and Xenopus (xIP3R) Ins(1,4,5)P3 receptors. (B) Ins(1,4,5)P3R expression levels in the different cell lines. Different cell lines were generated using the DT40 cell line where all three Ins(1,4,5)P3R isoform are deleted (KO). The 3KO cells were electroporated with the linearized plasmid for either the T930A or T930E mutants. The plasmid carrying the mutants contains G418 resistance. Stable cell lines for each mutant were established using G418 selection. WT: DT40 cell line expressing only the wild-type Ins(1,4,5)P3 receptor (type 1); T930A and T930E: expressing the respective mutant Ins(1,4,5)P3Rs. (C) Growth rates of the different cell lines are also equivalent. The same number of cells was plated at time zero and cells were counted at different time points under equivalent culturing conditions to determine whether the expression of the different mutants affects cell growth. Although the T930E mutant grew at a slightly slower rate the difference were not significant. (D) Resting Ca2+ levels and store Ca2+ content were measured in DT40 cells loaded with Fura-2 and incubated in Ca2+-free solution. Ionomycin was added as indicated to release Ca2+ from stores.
The Ins(1,4,5)P3 receptor requires both Ins(1,4,5)P3 and Ca2+ as co-agonists to gate open and release Ca2+ from stores. However receptor function is also modulated through phosphorylation by various kinases. The type 1 Ins(1,4,5)P3 receptor contains two canonical PKA phosphorylation sites S-1589 and S-1755.Citation6 PKA phosphorylation results in increased Ca2+ release activity.Citation7,Citation8 At the single channel level PKA phosphorylation increases the probability of opening (Po) of the Ins(1,4,5)P3 receptor resulting in bursting activity without affecting its Ca2+ dependence, arguing for an increase in the apparent sensitivity to Ins(1,4,5)P3.Citation8,Citation9
Although PKA phosphorylation is best understood, the Ins(1,4,5)P3 receptor has been shown to be phosphorylated and modulated by other kinases, including PKC, CaMKII and non-receptor tyrosine kinases.Citation10-Citation14 PKC phosphorylation of the Ins(1,4,5)P3 receptor leads to enhanced Ins(1,4,5)P3-dependent Ca2+ releaseCitation10,Citation11; and CaMKII phosphorylation modulates Ca2+ oscillations and inhibition of Ins(1,4,5)P3-dependent Ca2+ release.Citation12,Citation13
Ins(1,4,5)P3-dependent Ca2+ release is also modulated during the cell cycle with the most dramatic example at fertilization. Ca2+ release through Ins(1,4,5)P3 receptors is essential for egg activation and for mediating the egg to embryo transition in vertebrates.Citation15-Citation17 In Xenopus oocytes, Ins(1,4,5)P3-dependent Ca2+ release is sensitized during meiosis, and this sensitization is essential to generate the fertilization-specific Ca2+ transient that is required for egg activation at fertilization.Citation18-Citation20 In addition, we have recently shown that remodeling of the endoplasmic reticulum during oocyte meiosis contributes to Ins(1,4,5)P3 receptor sensitization through a process termed “geometric sensitization”Citation21. Geometric sensitization is due to ER remodeling during meiosis resulting in the formation of large ER patches enriched in Ins(1,4,5)P3 receptors. Ins(1,4,5)P3 receptors that localize to these ER patches display increased sensitivity to Ins(1,4,5)P3 as compared with their counterparts in the reticular ER.Citation21
We have also argued that phosphorylation by cell cycle kinases is involved in Ins(1,4,5)P3 receptor sensitization during meiosis.Citation22 Xenopus oocytes express the SII- variant of the type 1 Ins(1,4,5)P3 receptor, and the receptor is phosphorylated at both PKA sites throughout oocyte maturation.Citation22 During meiosis the Ins(1,4,5)P3 receptor is specifically phosphorylated on three additional residues T931, T1136 and T1145.Citation22 Both T931 and T1136 localize to consensus sites for proline-directed kinases such as MAPK and Cdk1. Furthermore, activation of the MAPK cascade or Cdk1 sensitizes Ins(1,4,5)P3-dependent Ca2+ release in Xenopus oocytes.Citation22 In fact phosphorylation of T1136 requires Cdk1 activity.Citation22 All three residues, T-931, T-1136 and S-1145, localize within the coupling domain and correspond to T-930, T-1140 and S-1152 of the rat SI, SII, SIII- isoform. The Ins(1,4,5)P3 receptor is also phosphorylated during mitosis.Citation23,Citation24
Here we test the role of phosphorylation at T-930 of the rat Ins(1,4,5)P3 receptor (SI, SII, SIII- splice variant). We focus on T-930 because it is specifically phosphorylated during meiosis and because the sequence surrounding this residue is conserved among vertebrates (). We show that a phosphomimetic mutation at T-930 (T930E) inhibits Ca2+ release through the Ins(1,4,5)P3 receptor. Furthermore, Ins(1,4,5)P3-binding affinity of the T930E mutant is significantly decreased compared with the wild-type or T930A mutant. These data show that Ins(1,4,5)P3 receptor phosphorylation at T-930, a site within the coupling domain, decreases Ins(1,4,5)P3 binding affinity of the receptor resulting in decreased Ins(1,4,5)P3-dependent Ca2+ release.
Results
To investigate the role of phosphorylation at T-930 on Ins(1,4,5)P3 receptor function, we engineered two mutants one with an alanine substitution at T-930 (T930A), and the other with a phosphomimetic mutation where the threonine is replaced by a glutamic acid (T930E). We chose glutamic acid as its side chain closely resembles that of a phosphorylated negatively charged threonine side chain. As such the T930E mutant is expected to mimic the behavior of the Ins(1,4,5)P3 receptor phosphorylated at T-930. In contrast, the alanine substitution at T930 serves as a control and provides a mutant that is not phosphorylatable at this residue. Furthermore, it also controls for the structural need for a threonine at this position.
The ubiquitous distribution of the Ins(1,4,5)P3 receptor makes it challenging to study the effect of different mutants on Ins(1,4,5)P3 receptor function because of the background signal due to endogenous Ins(1,4,5)P3 receptors in most cells. Therefore to study the functional consequences of phosphorylation at T-930, we used the DT40 chicken lymphocyte cell line because of the existence of a DT40 line (3KO), where all three Ins(1,4,5)P3 receptor genes (type 1, 2, and 3) have been knocked out. This provides a clean background to analyze the function of different expressed Ins(1,4,5)P3 receptor mutants. Using the 3KO line as the parental line, we generated stable DT40 cell lines expressing the wild-type rat Ins(1,4,5)P3 receptor, the T930A or the T930E mutants. Following the establishment of the cell lines we amplified and sequence confirmed that they are in fact expressing the relevant Ins(1,4,5)P3 receptor mutant.
Western blot analysis shows that the three different cell lines express the Ins(1,4,5)P3 receptor at equivalent levels, whereas no immuno-reactivity is detected in the 3KO line (). The growth rate of the different DT40 cell lines were equivalent, arguing that expression of the different mutants did not have a dramatic effect on cell survival or ability to replicate (). Finally, both resting Ca2+ levels and Ca2+ store content were similar among the different cell lines (), showing that expression of the T930E or T930A mutant does not alter Ca2+ homeostasis significantly at rest.
To test the sensitivity of the different Ins(1,4,5)P3 receptor mutants, we developed an assay to measure Ca2+ release from intracellular stores in situ in response to an Ins(1,4,5)P3 dose response. For these experiments DT40 cells were loaded with caged-Ins(1,4,5)P3 and Ins(1,4,5)P3 was uncaged for different durations as indicated in . Cells expressing wild-type (WT) Ins(1,4,5)P3 receptor respond with an exponential increase in Ca2+ release along the Ins(1,4,5)P3 uncaging dose response (). This assay provides a direct test of Ins(1,4,5)P3 receptor function because Ins(1,4,5)P3 produced following uncaging binds to and gates the Ins(1,4,5)P3 receptor without invoking additional intermediaries.
Figure 2. The T930E mutant exhibits decrease Ins(1,4,5)P3-dependent Ca2+ release sensitivity. DT40 cells were loaded with a Ca2+ dye and caged-Ins(1,4,5)P3 and exposed to different UV uncaging pulse durations to produce a dose response of Ins(1,4,5)P3 intracellulary. The level of Ca2+ release following the uncaging pulses was used as a measure of the sensitivity of the Ins(1,4,5)P3 receptor mutants as compared with the wild-type receptor.
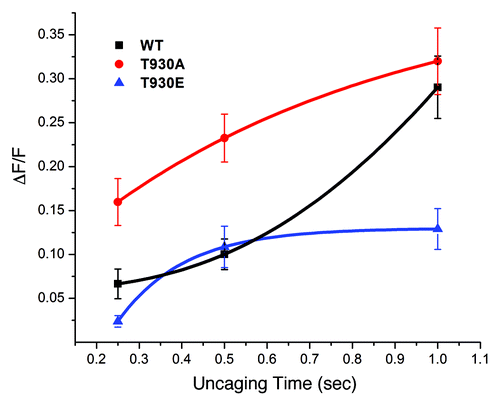
Surprisingly in cells expressing the T930A mutant Ins(1,4,5)P3-dependent Ca2+ release is sensitized compared with the wild-type receptor (). The T930A mutant receptor responds with increased Ca2+ release at lower Ins(1,4,5)P3 concentrations although the maximum amount of Ca2+ release is comparable to the wild-type receptor (). This sensitization is unlikely to be due to higher expression levels of the T930A mutant as compared with WT, because the different cell lines express equivalent levels of Ins(1,4,5)P3 receptors () and because the maximal Ca2+ release is similar between the two cell lines ().
In contrast, cells expressing the T930E mutant exhibit the opposite phenotype of decreased Ca2+ release in response to Ins(1,4,5)P3 uncaging (). Interestingly, even at high levels of Ins(1,4,5)P3 these cells release significantly smaller levels of Ca2+ (). This is not due to decreased store Ca2+ content, since T930E expressing cells have a similar store Ca2+ load as cells expressing the wild-type receptor or the T930A mutant (). In contrast cells expressing the T930A mutant or the wild-type receptor produce significantly higher Ca2+ release levels at long uncaging durations (). Because the T930E cell line expresses equivalent amount of Ins(1,4,5)P3 receptors, these results argue that either the affinity of the T930E mutant to Ins(1,4,5)P3 is low or that gating/permeation or Ca2+ dependence of the receptor are affected by the T930E mutation leading to short lived or small Ca2+ release.
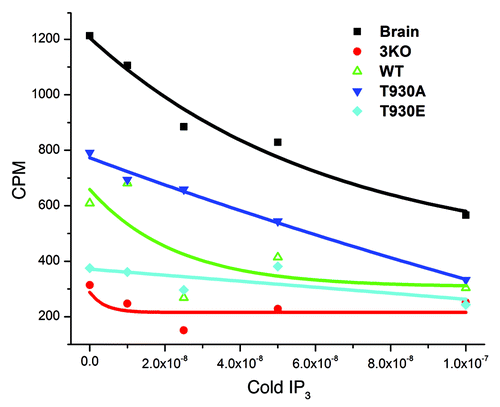
To further investigate which of these mechanisms is affected in the T930E mutant, we directly measured the Ins(1,4,5)P3 binding affinity of the different mutants in microsomes derived from the stable DT40 cell lines (). Microsomes from brain, which is enriched in Ins(1,4,5)P3 receptors, were used as a positive control and showed significant Ins(1,4,5)P3-binding that was competed effectively by cold Ins(1,4,5)P3 (). Ins(1,4,5)P3 binding to microsomes isolated for the DT40 cell lines expressing the wild-type Ins(1,4,5)P3 receptor showed lower binding than brain microsomes, which was also effectively competed away using cold Ins(1,4,5)P3 (). Consistent with the Ins(1,4,5)P3 uncaging data (), microsomes from cells expressing the T930A mutant exhibited higher Ins(1,4,5)P3 binding affinity than their wild-type counterparts, whereas Ins(1,4,5)P3 binding to T930E microsomes was hardly detectable and only slightly above the background binding observed in the 3KO cells (). These data show that the behavior of the T930A and T930E mutants in situ can be explained solely based on their Ins(1,4,5)P3 binding affinity without the need to invoke alterations to Ca2+ dependence, gating or permeation of the receptor. Our results further show that phosphorylation at T930 leads to a significant decrease in Ins(1,4,5)P3 binding affinity.
Figure 3. The T930E mutation decreases Ins(1,4,5)P3 binding affinity. Microsomal preparations from brain as a positive control and from the different DT40 cell lines as indicated were subjected to the Ins(1,4,5)P3 binding assay to measure the binding affinity of Ins(1,4,5)P3 receptor mutants. Briefly lysates were allowed to bind radioactively (3H) labeled Ins(1,4,5)P3 and the label competed with cold Ins(1,4,5)P3. The competitive assay provides a relative comparative measure Ins(1,4,5)P3R affinity in the different mutants as compared with the wild type receptor (WT). The negative control was the 3KO cell line which does not express any of the three Ins(1,4,5)P3 receptor isoforms and as such provides non-specific background binding.
Discussion
Using stable DT40 cell lines expressing different Ins(1,4,5)P3 receptor mutants we show that phosphorylation of the type 1 receptor at T930 is likely to decrease Ins(1,4,5)P3-binding affinity. As discussed above the Ins(1,4,5)P3 receptor is targeted by various kinases and in most cases phosphorylation results in increased sensitization of Ins(1,4,5)P3-dependent Ca2+ release.Citation1 In contrast, a phosphomimetic mutation at T930 results in decreased Ins(1,4,5)P3 binding affinity and reduced Ins(1,4,5)P3-dependent Ca2+ release. The T930 residue localizes within the coupling domain and is distant from the Ins(1,4,5)P3 binding domain in the linear sequence of the Ins(1,4,5)P3 receptor. This brings into question the ability of phosphorylation at this site to modulate Ins(1,4,5)P3 binding affinity. However, the available structure of the Ins(1,4,5)P3 receptor obtained from cryo-EM studies shows that the coupling domain lies below the Ins(1,4,5)P3 binding domain in the 3-D structure of the receptor.Citation25 Hence phosphorylation at T-930 could modulate Ins(1,4,5)P3 binding if this residue interacts with the Ins(1,4,5)P3-binding and/or suppressor domains. Alternatively, phosphorylation at T930 could alter the conformation of the coupling domain in close proximity of the Ins(1,4,5)P3-binding domain thus modulating its ability to bind Ins(1,4,5)P3. One argument supporting this possibility is the sensitization of Ins(1,4,5)P3-dependent Ca2+ release observed in the cells expressing the T930A mutant. This sensitization argues that structural/sequence modifications within this region of the coupling domain affect Ins(1,4,5)P3 receptor sensitivity.
The Ins(1,4,5)P3 receptor is phosphorylated at T930 during meiosis. Furthermore, Ins(1,4,5)P3-dependent Ca2+ release is highly sensitized during meiosis and this sensitization is critical for egg activation and the egg-to-embryo transition at fertilization. This creates a conundrum, since the phosphomimetic T930E mutation decreases Ins(1,4,5)P3 binding affinity and as such reduces the sensitivity of Ins(1,4,5)P3-dependent Ca2+ release. This is quite interesting as it implicates cross talk among different phosphorylation sites within the Ins(1,4,5)P3 receptor to define its biological function. During meiosis the Xenopus Ins(1,4,5)P3 receptor is also phosphorylated at two additional residues that match the consensus for proline-directed kinases, T-1136 and S-1145. So it is possible that the combinatorial phosphorylation at all three residues sensitizes Ins(1,4,5)P3-dependent Ca2+ release. Another formal possibility is that the rat receptor used in this study behaves differently in response to phosphorylation at T-930 as compared with the Xenopus receptor. However, given the high degree of sequence conservation between the two receptors (89% identity) this possibility seems remote. In addition, because of the high levels of endogenous PKA activity in oocytes, the Ins(1,4,5)P3 receptor is also phosphorylated at the consensus PKA sites during maturation.Citation22 Interestingly, all these residues localize within the coupling domain, arguing that combinatorial phosphorylation within this domain may differentially affect Ins(1,4,5)P3 binding affinity and the function of the Ins(1,4,5)P3 receptor.
Materials and Methods
Clones and cell culture
Plasmid pcDNA3 containing the rat Ins(1,4,5)P3 receptor (pcDNA3 rIP3R) was a kind gift from Suresh Joseph. Mutations in the coding region of the Ins(1,4,5)P3 receptor were generated using Quick Change II Site-Directed Mutagenesis kits (Strategene) to change the bases “ACT” to “GCT” in the coding DNA strand resulting in an amino acid substitution of threonine (T930) with alanine (T930A). Likewise, “ACT” was replaced with “GAA” resulting in the substitution of amino acid threonine (T930) with glutamic acid (T930E). Mutations were confirmed by sequence analysis.
To generate DT40 cell lines expressing the mutated IP3R, linearized pcDNA3 T930A and pcDNA3 T930E plasmids were electroporated into a DT40–3KO cell line where all three Ins(1,4,5)P3 receptor isoforms are deleted. Stable cell lines were established using G418 selection.
Western blot analysis
Lysates from ~5 × 107 DT40 cells expressing wild-type rIP3R, T930A, T930E, or from the 3KO cell line were separated on denaturing NuPAGE 3–8% Tris-Acetate gradient gels (Invitrogen). Primary rabbit anti-IP3R (T443) antibody and secondary goat anti-rabbit-HRP antibody (Jackson Immunoresearch) at 1:1000 and 1:7500 dilution, respectively, in 1% Hammerstein casein, 2% BSA were used for western analysis. Ins(1,4,5)P3 receptor protein bands were detected using ECL-Plus (Amersham) detection reagent.
Ins(1,4,5)P3 binding assay
Microsomes were prepared from rabbit cerebellum by homogenizing 1g of tissue in 12 ml of E Buffer (20 mM TRIS-HCl pH 8.3, 10 mM KCl, 1mM EDTA, 1 mM DTT, Cocktail inhibitor III (Calbiochem) and 1 mM PMSF) using a Polytron homogenizer. The lysate was centrifuged at 1000 xg for 15 min at 4°C and the resulting supernatant was transferred to a microfuge tube and placed on ice. The pellet was suspended in 3 ml of E buffer and centrifuged again at 1000x g. The supernatants were combined and centrifuged at 2000 xg for 15 min. The resulting supernatant was then centrifuged at 105,000 xg for 30 min. The pellet containing the microsomes was suspended in E buffer and the protein concentration was determined using a BioRad protein assay.
DT40 cells were collected by centrifugation at 1,500 xg for 15 min. The cell pellets were washed once in E Buffer then centrifuged at 1,500 xg for 15 min. The cells were lysed with 20 strokes in a glass Dounce homogenizer (Pyrex 7727-15) in 5 ml of E buffer. Cell lysates were centrifuged at 1,500 xg for 15 min. The supernatants were transferred to polyallomer ultracentrifuge tubes (Beckman #326814) and centrifuged at 100,000 xg for 30 min at 4°C. The pellets were suspended in E buffer and protein concentrations were determined using a BioRad protein assay.
Ins(1,4,5)P3 binding assays were performed using microsome preparations from rabbit cerebellum and DT40 cells expressing wild-type Ins(1,4,5)P3 receptor, T930A, T930E, and from the 3KO DT40 cells. Each reaction contained 120 µg of brain or 1 mg of DT40 microsomes. The reaction mix contained 15 nM [3H]Ins(1,4,5)P3 (5 nM [3H]Ins(1,4,5)P3/µl NET911, 0.005mCi, 21.4Ci/mmol); various concentrations ranging from 0–2,000 µM of cold Ins(1,4,5)P3 (10 mM Ins(1,4,5)P3, Invitrogen), Ins(1,4,5)P3 binding buffer (50 mMTRIS-HCl, pH 8.3, 1 mM EDTA, 1 mM DTT, 0.1 M KCl), EDS buffer (20 mM TRIS-HCl, pH 8.3, 20 mM NaCl, 10 mM KCl, 1mM EDTA, 1 mM DTT, 10% sucrose plus protease inhibitors) in a final volume of 500 µl. The reaction mixtures were incubated on ice for 10 min and then 5 µl of γ-globulin and 5 µl of glycogen were added and incubated for additional 5 min. The mixtures were centrifuge at 20,000 xg for 15 min, the supernatants were aspirated, and the pellets were solubilized in Solubilization buffer (0.625 M TRIS-HCl, pH 7.5, 5% 2-mercaptoethanol, 2% SDS) and the amount of 3H was determined using a scintillation counter.
Ins(1,4,5)P3 uncaging n DT40 cells
Measurements of intracellular free calcium concentration ([Ca2+]i) were made using the fluorescent indicator Ca2+-green-AM. Cells (approximately 2.5 × 106) were loaded for 45 min in growth medium at 40° containing 5 μM ci-IP3, 2 μM Ca2+-green-AM and 0.0025% Pluronic F-127. Cells were washed three times and re-suspended at 4°C in HBSS (in mM: 137 NaCl, 5.4 KCl, 0.25 Na2HPO4, 0.44 KH2PO4, 1.3 CaCl2, 1.0 MgSO4, 4.2 NaHCO3, 5.5 D-glucose, 10 Hepes pH 7.4) to a final density of 106 cells ml−1 and kept on ice for up to 1 h. Cells were then pre-warmed to 40° for 15 min. Fluorescence of the stirred cell suspension was then measured by emission at 520 nm while uncaging of ci-Ins(1,4,5)P3 was performed at 330 ± 40 nm. Removal of all residual extracellular calcium was achieved by addition of 3 mM EGTA at the beginning of fluorescence measurements.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by a grant from the National Institutes of Health, GM61829, and by a grant from the Qatar National Research Fund (QNRF), NPRP 08-395-3-088. R01-DK054568 to D.I.Y. The statements made herein are solely the responsibility of the authors. Additional support for the authors comes from Biomedical Research Program funds at Weill Cornell Medical College in Qatar, a program funded by Qatar Foundation.
References
- Foskett JK, White C, Cheung KH, Mak DO. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev 2007; 87:593 - 658; http://dx.doi.org/10.1152/physrev.00035.2006; PMID: 17429043
- Patterson RL, Boehning D, Snyder SH. Inositol 1,4,5-trisphosphate receptors as signal integrators. Annu Rev Biochem 2004; 73:437 - 65; http://dx.doi.org/10.1146/annurev.biochem.73.071403.161303; PMID: 15189149
- Mikoshiba K. The InsP3 receptor and intracellular Ca2+ signalling. Curr Opin Neurol 1997; 7:339 - 45; http://dx.doi.org/10.1016/S0959-4388(97)80061-X
- Bezprozvanny I, Watras J, Ehrlich BE. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature 1991; 351:751 - 4; http://dx.doi.org/10.1038/351751a0; PMID: 1648178
- Mak DO, McBride S, Foskett JK. Inositol 1,4,5-trisphosphate [correction of tris-phosphate] activation of inositol trisphosphate [correction of tris-phosphate] receptor Ca2+ channel by ligand tuning of Ca2+ inhibition. Proc Natl Acad Sci U S A 1998; 95:15821 - 5; http://dx.doi.org/10.1073/pnas.95.26.15821; PMID: 9861054
- Ferris CD, Cameron AM, Bredt DS, Huganir RL, Snyder SH. Inositol 1,4,5-trisphosphate receptor is phosphorylated by cyclic AMP-dependent protein kinase at serines 1755 and 1589. Biochem Biophys Res Commun 1991; 175:192 - 8; http://dx.doi.org/10.1016/S0006-291X(05)81219-7; PMID: 1847804
- Volpe P, Alderson-Lang BH. Regulation of inositol 1,4,5-trisphosphate-induced Ca2+ release. II. Effect of cAMP-dependent protein kinase. Am J Physiol 1990; 258:C1086 - 91; PMID: 2163198
- Wagner LE 2nd, Joseph SK, Yule DI. Regulation of single inositol 1,4,5-trisphosphate receptor channel activity by protein kinase A phosphorylation. J Physiol 2008; 586:3577 - 96; http://dx.doi.org/10.1113/jphysiol.2008.152314; PMID: 18535093
- Tang TS, Tu H, Wang Z, Bezprozvanny I. Modulation of type 1 inositol (1,4,5)-trisphosphate receptor function by protein kinase a and protein phosphatase 1alpha. J Neurosci 2003; 23:403 - 15; PMID: 12533600
- Ferris CD, Huganir RL, Bredt DS, Cameron AM, Snyder SH. Inositol trisphosphate receptor: phosphorylation by protein kinase C and calcium calmodulin-dependent protein kinases in reconstituted lipid vesicles. Proc Natl Acad Sci U S A 1991; 88:2232 - 5; http://dx.doi.org/10.1073/pnas.88.6.2232; PMID: 1848697
- Matter N, Ritz MF, Freyermuth S, Rogue P, Malviya AN. Stimulation of nuclear protein kinase C leads to phosphorylation of nuclear inositol 1,4,5-trisphosphate receptor and accelerated calcium release by inositol 1,4,5-trisphosphate from isolated rat liver nuclei. J Biol Chem 1993; 268:732 - 6; PMID: 8380171
- Zhu DM, Tekle E, Chock PB, Huang CY. Reversible phosphorylation as a controlling factor for sustaining calcium oscillations in HeLa cells: Involvement of calmodulin-dependent kinase II and a calyculin A-inhibitable phosphatase. Biochemistry 1996; 35:7214 - 23; http://dx.doi.org/10.1021/bi952471h; PMID: 8679550
- Matifat F, Hague F, Brûlé G, Collin T. Regulation of InsP3-mediated Ca2+ release by CaMKII in Xenopus oocytes. Pflugers Arch 2001; 441:796 - 801; http://dx.doi.org/10.1007/s004240000479; PMID: 11316263
- Jayaraman T, Ondrias K, Ondriasová E, Marks AR. Regulation of the inositol 1,4,5-trisphosphate receptor by tyrosine phosphorylation. Science 1996; 272:1492 - 4; http://dx.doi.org/10.1126/science.272.5267.1492; PMID: 8633244
- Stricker SA. Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev Biol 1999; 211:157 - 76; http://dx.doi.org/10.1006/dbio.1999.9340; PMID: 10395780
- Miyazaki S, Shirakawa H, Nakada K, Honda Y. Essential role of the inositol 1,4,5-trisphosphate receptor/Ca2+ release channel in Ca2+ waves and Ca2+ oscillations at fertilization of mammalian eggs. Dev Biol 1993; 158:62 - 78; http://dx.doi.org/10.1006/dbio.1993.1168; PMID: 8392472
- Machaca K. Ca2+ signaling differentiation during oocyte maturation. J Cell Physiol 2007; 213:331 - 40; http://dx.doi.org/10.1002/jcp.21194; PMID: 17620315
- Terasaki M, Runft LL, Hand AR. Changes in organization of the endoplasmic reticulum during Xenopus oocyte maturation and activation. Mol Biol Cell 2001; 12:1103 - 16; PMID: 11294910
- Machaca K. Increased sensitivity and clustering of elementary Ca2+ release events during oocyte maturation. Dev Biol 2004; 275:170 - 82; http://dx.doi.org/10.1016/j.ydbio.2004.08.004; PMID: 15464580
- Ullah G, Jung P, Machaca K. Modeling Ca2+ signaling differentiation during oocyte maturation. Cell Calcium 2007; 42:556 - 64; http://dx.doi.org/10.1016/j.ceca.2007.01.010; PMID: 17349690
- Sun L, Yu F, Ullah A, Hubrack S, Daalis A, Jung P, et al. Endoplasmic reticulum remodeling tunes IP₃-dependent Ca²+ release sensitivity. PLoS One 2011; 6:e27928; http://dx.doi.org/10.1371/journal.pone.0027928; PMID: 22140486
- Sun L, Haun S, Jones RC, Edmondson RD, Machaca K. Kinase-dependent regulation of IP3-dependent Ca2+ release during oocyte maturation. J Biol Chem 2009; 284:20184 - 96; http://dx.doi.org/10.1074/jbc.M109.004515; PMID: 19473987
- Malathi K, Li X, Krizanova O, Ondrias K, Sperber K, Ablamunits V, et al. Cdc2/cyclin B1 interacts with and modulates inositol 1,4,5-trisphosphate receptor (type 1) functions. J Immunol 2005; 175:6205 - 10; PMID: 16237118
- Lee B, Vermassen E, Yoon SY, Vanderheyden V, Ito J, Alfandari D, et al. Phosphorylation of IP3R1 and the regulation of [Ca2+]i responses at fertilization: a role for the MAP kinase pathway. Development 2006; 133:4355 - 65; http://dx.doi.org/10.1242/dev.02624; PMID: 17038520
- Ludtke SJ, Tran TP, Ngo QT, Moiseenkova-Bell VY, Chiu W, Serysheva II. Flexible architecture of IP3R1 by Cryo-EM. Structure 2011; 19:1192 - 9; PMID: 21827954