Abstract
The analysis of genomes suggests that horizontal transfers are frequent phenomena. In eukaryotes these transfer often involve transposable elements and can be detected by sequence analysis or phylogenetic reconstruction. Nevertheless, the dynamics of transposable elements and reticulation in species history, especially in plants, can sometimes be misleading. While the horizontal transfer of transposable elements is well documented in animals, only two cases have been described in plants despite the abundance of these elements in plant genomes. The study of horizontal transfers of transposable elements in plants represents a new challenge to understand their impact on genomic diversity and consequently on the process of adaptation to their environment.
Introduction
The transmission of genetic information among living organisms usually occurs vertically by sexual or asexual reproduction from parents to their progeny. In some cases, the genetic material can be transmitted between reproductively isolated species, a mechanism referred to as Horizontal Transfer (HT).Citation1,Citation2 In prokaryotes, HTs have occurred so frequently that they seem to be the main cause of phylogenetics incongruences. They are therefore considered to play a major role in the evolution and speciation in prokaryotes.Citation3 The recent increase of genomic data has allowed to conduct extensive surveys of some eukaryotic genomes and confirmed that HTs, initially considered as rare phenomena, are also quite common and recurrent in these organisms. So far, most of the documented cases of HT concern genes. Moreover, Horizontal Gene Transfers (HGTs) appear to involve organisms independently from their phylogenetic relatedness. There are cases of HGT between species of the same genus,Citation4 between species of different genusCitation5 and even between organisms belonging to distinct kingdoms,Citation6,Citation7 especially in the context of symbioses.Citation8 Furthermore Holmgren et al.,Citation9 detected transfers among human cells where DNA coming from apoptotic bodies can be captured by surrounding cells. This, therefore, suggests that such transfer could therefore play a major role in evolution and adaptation of organisms to their environment.Citation10
Transposable Elements (TEs) are DNA sequences which are naturally mobile in their host genome. They are divided into two classes according to their mode of transposition.Citation11 Class I, or retrotransposons, transpose via a “copy and paste” mechanism. This involves a RNA intermediate, which is reverse transcribed and integrated into the genome as a duplicate copy. Class I elements can therefore increase their copy number while active. Class II, or transposons, transpose through a “cut and paste” mechanism. These elements are excised and directly reintegrated elsewhere in the genome. Because TEs (whether they belong to Class I or Class II) exist as a free molecule in the cell at least at one point in their transposition cycle, one could envisage that they may be more prone to HTs than any other genomic sequence, given that they could be in contact with a vector that could mobilize them to another organism.Citation12 In addition, some elements contain envelope-like coding domains, which make them structurally similar to retroviruses.Citation13,Citation14 However, even if TEs are the main components of plant genomes,Citation15 only few cases of TEs HT have been described,Citation16 which may be an indication that they are a rare phenomenon. Alternatively, the scarcity of cases of TE HTs in the literature could only reflect the technical difficulties raised by their detection and correct characterization. In this review, we will therefore first describe the different approaches that have been used so far to detect HTs in eukaryotes. We will then discuss the importance of HTs in TE biology and more generally in plant genome evolution as well as the putative mechanisms involved in such process.
Detection of Horizontal Transfer of Transposable Elements
Three approaches have been used to detect HT. The most common, used in prokaryotes, is based on a search of bias in nucleotide composition. This is possible if the portion of transferred DNA is in a genetic context, which is different from that of the former host: since each of the two organisms has experienced unique selective constraints, they may harbor different genomic signatures and different biases in codon usage or nucleotide composition.Citation17 These methods were used to develop accurate algorithms,Citation18 but the two organisms have to be divergent enough to display such biases.
The other approach is based on phylogenetic reconstruction. HTs can generate phylogenetic incongruence: the topology of the phylogenetic tree obtained from the horizontally transferred sequence differs significantly from that obtained using classical genes.Citation19 This approach requires a statistical validation to highlight the discordances: these are (from the quickest but less accurate to the more time consuming but precise) the Kishino and Hasegawa test (KH),Citation20 the Shimodaira and Hasegawa test (SH),Citation21 the SOWH test from Goldman et al.,Citation22 or the more robust Expected Likelihood Weights test (ELW) of Strimmer and Rambaut.Citation23
A third method is specific to TEs and consists in using the Long Terminal Repeats (LTR)-retrotransposons to detect HTs. These elements mutate faster than gene,Citation24,Citation25 and are rapidly deleted through unequal and illegitimate recombination, which leads to their fast turn-over in eukaryote genomes.Citation24,Citation26 It is possible to test whether the divergence between LTR-retrotransposon sequences of two distinct species is congruent with their radiation and predict whether vertical or horizontal transmission of the element occurred. Furthermore, the divergence between the two LTR of the retrotransposon can be used to estimate the age of the insertion of the element and therefore infer the age of the HT.Citation27
Limits of the Detection Methods
The three methods of HT detection described above exploit the incongruence between the sequence identity among TE-related sequences and the overall divergence of the genomes of the taxa involved in the transfer. However, two other mechanisms may lead to similar observations and should therefore be taken into account very carefully before any conclusion regarding horizontal transfer could be made. These are the transfer via interspecific introgressions and the domestication of TE-related sequences.
Introgressions involve the transfer of a DNA sequence from one species to the other through recurrent backcrossing of an interspecific hybrid with one of its parents. This phenomenon is very common in plantsCitation19 and can also occur in animals.Citation28 If introgressed genomic regions harbor TEs, then the transferred DNA segment generates incongruence among data sets and may present signatures or composition different from those of the host in which it is transferred. The only way to ascertain whether this DNA comes from HT or introgression is to check the flanking regions of the putative transferred copy. Introgression usually conserves synteny (i.e., the homology of the TE and of both flanking genomic sequences) whereas HT does not (because only the transposed TE will exhibit a strong sequence identity with an element of the other parent). In addition, as introgression can only occur through sexual reproduction, it first requires the spontaneous hybridization of two sympatric species. These species should thus be close enough phylogenetically to allow such crosses to occur. Therefore, introgressions can only concern species belonging to the same genus, or eventually to two different genera, albeit closely related.
The domestication of TEs by their host genome is defined as the co-option of TE-encoded protein or domain into functional host protein.Citation29 In such case, these TE-related sequence can be under strong selection because they control an essential biological function of the host and therefore evolve at a much lower rate than other TEs. For example, in human, two genes, Apolipoprotein-C1 (Apo-C1) and Endothelin B Receptor (EBR) use the LTR of an endogenous retrovirus as alternative promoters.Citation30 While the insertions are 20–30 and 30–40 My old for Apo-C and EBR respectively, the LTR of both loci are highly conserved, showing 88% of sequence identity. More surprising are the 223 bp of the uc.338 element, which are perfectly conserved between human, rat and mouse. This element also exhibits a high sequence identity with a putative SINE of Coelacanth, whereas both lineages diverged 400 MYA.Citation31 This strong selection, leading to minor variations in the TE sequence causes to underestimate the dates of the insertions in both organisms and therefore mimics a HT. Furthermore, mouse enhancer assays show that one copy of the uc.338 element could act as enhancer of a neurodevelopmental gene. These two examples show that TEs can gain function and be submitted to strong selective forces. This could therefore explain TEs sequence conservation between species. However, most examples of domestication concern only some domain of TEs and more particularly the transposase encoding sequence.Citation29 There is to our knowledge no example of domestication that concerns the full length of the element, including non-functional regions. In such case, the HT hypothesis is considered to be more parsimonious to explain the high conservation of elements throughout its sequence and among different species.
Extent of Horizontal Transfers of Transposable Elements
In animals, several cases have demonstrated that horizontal gene transfers (HGT) occurred frequentlyCitation8,Citation32 and most of them involve TEs. The HT of the P-element in Drosophila represents one of the best described cases of such event.Citation4,Citation33,Citation34 This element present originally in D. willistoni transferred to D. melanogaster spread rapidly through the populations of this species over the last fifty years.Citation33 The HT of this element has been reported to have occurred four times within the Drosophila genus.Citation35 Drosophila is a good model to study HTs of TEs since many examples of such transfers have been described within this genus (e.g., MinosCitation36 and a copia elementCitation37) and between this genus and others (e.g., MarinerCitation38). In animals many other cases of HTs of TEs have been suspected, e.g., crustacean genomes (MarinerCitation39), among fish (SINE family,Citation40 Rex1Citation41), between reptiles and mammalsCitation12 and between mammals and hookworms (bandit transposonCitation42).
Although HTs have been well documented in plants,Citation5,Citation43–Citation46 only two cases of transfer of TEs have been described. The first one concerns the HT of a Mu-like (Class II) between foxtail millet and rice.Citation16 Because both species diverged more than 50 MY ago, these homologous sequences should not present homology unless they were subjected to selection or to horizontal transfer. The authors showed that the whole element displays no trace of selection and therefore concluded in favor of a horizontal transmission of the element. The second case showed that an LTR-retrotransposon, RIRE1 been involved at least in seven HTs among the genus OryzaCitation27 was based on the high conservation of RIRE1 sequences between distant and reproductively isolated wild rice species. The authors also demonstrated that the transferred copy was still active after the HT and that RIRE1 now composes a large part of some genomes of the genus.Citation47 This result is in accordance with the putative role of horizontal transfer in genome evolution. These two examples provide evidence that in plants, HT of TEs can occur like in animals. The fact that it occurs between distantly related species suggests that it is a common mechanism in the genome evolution of both plants and animals.
Mechanisms of Transposable Elements Horizontal Transfers
Three hypotheses have been proposed to explain TEs HTs. The first one is a direct transfer of DNA between two plants that are in close contact. Indeed, in some cases, parasitic angiosperms are involved in HGT and demonstrate that direct plant-to-plant HGT can occur.Citation44,Citation45 This plant-to-plant mechanism has been proposed to explain the massive HGT from land plant to Amborella trichopoda.Citation43 This mechanism is likely to occur for any DNA sequence, gene or TE.
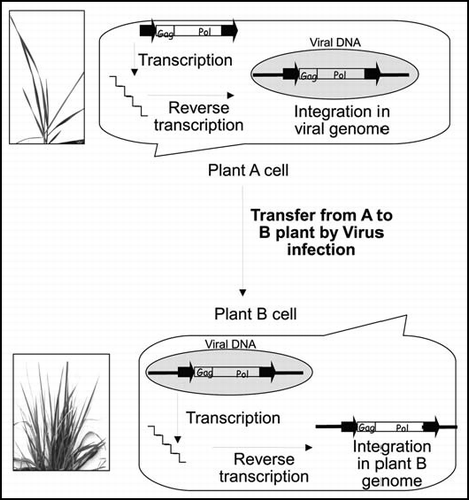
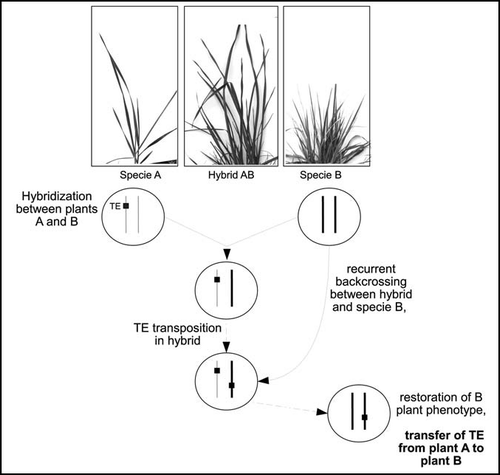
However, the two well-documented examples of HT of TEs in plants are more in favor of a vector-mediated transfer than of a direct plant-to-plant transfer. One could argue that plant-to-plant transfer could be facilitated in plants because of the occurrence of interspecific crosses. Horizontal transposition during such crosses, that is to say transposition of an element from one genome to another within the hybrid (), could facilitate the transfer of TE from one species to the other. The interspecific hybrids could constitute a bridge between species. Nevertheless, this second hypothesis could not easily explain the transposon HT between Setaria and riceCitation16 mainly because both species have diverged long time ago and spontaneous Setaria x Oryza hybrids are not likely to occur in wild populations. In the case of the HT of RIRE1 in the genus Oryza,Citation27 the high frequency of this transfer (seven species involved in the HT) does not fit well with the hypothesis of direct plant-to-plant transfer since the species do not occupy the same habitats. It is also not parsimonious to explain the seven HTs by the occurrence of seven independent interspecific hybridizations.
A third hypothesis could be that the transfers can be mediated by a vector (). In animals, several vectors are supposed to be involved in HT such as bacteria and fungus.Citation8,Citation32 It has been demonstrated that viruses can also be transitional hosts for the TE, as it has been proposed by Piskurek and Okada.Citation12 The authors show that a SINE element transposed from the genome of a reptile to a viral vector (poxvirus) and then probably from the vector to mammals. The authors use the term of horizontal transposition in reference to the mechanisms by which the TE is activated in the snake's cell and integrated into the genome of the virus. In plants, no vector involved in TEs HTs has been clearly identified but one could expect that horizontal transposition could occur in a similar way. This putative common mechanism could explain multiple and massive horizontal transfers of TE described both in animals (e.g., the DrosophilaCitation4) and in plants (e.g., OryzaCitation27). Moreover, insects such as sapsucker should be considered as potential vectors for HT in plants but this hypothesis is purely speculative and needs to be experimentally ascertained.
Conclusions
The understanding of mechanisms involved in HT represents a new challenge for the study of TEs evolution between species. The increase of available genomic sequences will provide more opportunities to confirm their occurrence in plants as well as to understand the mechanisms involved in these processes. Providing that these surveys demonstrate the high frequency of TEs HTs in plants, it would suggest that HTs are part of the natural “life cycle” of TEs. Such process could allow TEs to invade and survive in new genomes and hence to escape deletion and silencing in a given species.
The impact of TEs horizontal transfer on genome structure and gene expression has to be also investigated. KidwellCitation48 suggested that TEs HT could lead to adaptive evolution by generating variability. By their ability to modulate and control gene expression depending on their genomic insertion,Citation49 the transferred TE may play a major role in the adaptation of plants to their environment.
Abbreviations
| HTs | = | horizontal transfers |
| TEs | = | transposable elements |
| LTR | = | long terminal repeat |
Figures and Tables
Figure 1 Horizontal transposition: hybridization between species A and B is followed by TE activation in the hybrid. TE is this way transferred from sub-genome A to B. Recurrent backcrosses between the hybrid and species B lead to the restoration of plant B phenotype (presence of B chromosomes only) but harboring the transferred element.
References
- Syvanen M. Horizontal gene transfer: evidence and possible consequences. Annu Rev Genet 1994; 28:237 - 261
- Brown JR. Ancient horizontal gene transfer. Nat Rev Genet 2003; 4:121 - 132
- Lawrence JG. Gene transfer in bacteria: speciation without species?. Theor Popul Biol 2002; 61:449 - 460
- Silva JC, Kidwell MG. Horizontal transfer and selection in the evolution of P elements. Mol Biol Evol 2000; 17:1542 - 1557
- Won H, Renner S. Horizontal gene transfer from flowering plant to Gnetum. Proc Natl Acad Sci USA 2003; 100:10824 - 10829
- Veronico P, Jones J, Di Vito M, De Giorgi C. Horizontal transfer of a bacterial gene involved in polyglutamate biosynthesis to the plant parasitic nemathod Meloidogyne artiellia. FEBS Letts 2001; 508:470 - 474
- Intieri MC, Buatti M. The horizontal transfer of Agrobacterium rhizogenes genes and the evolution of the genus Nicotiana. Mol Phyl Evol 2001; 20:100 - 110
- Kondo N, Nikoh N, Ljichi N, Shimada M, Fukatsu T. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci USA 2002; 99:4280 - 4285
- Holmgren L, Szeles A, Rajnavölgyi E, Folkman J, Klein G, Ernberg I, Falk KI. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood 1999; 93:3956 - 3963
- Springael D, Top EM. Horizontal gene transfer and microbial adaptation to xenobiotics: new type of mobile genetic elements and lessons from ecological studies. Trens Microbiol 2004; 12:53 - 58
- Kasazhian HH. Mobile elements: drivers of genome evolution. Science 2004; 303:1626 - 1632
- Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptile to mammals. Proc Natl Acad Sci USA 2007; 104:12046 - 12051
- Laten HM, Havecker ER, Farmer LM, Voytas DF. Sire1, an endogenous retrovirus family from glycine max, is highly homogeneous and evolutionarily young. Mol Biol Evol 2003; 20:1222 - 1230
- Vicient CM, Kalendar R, Schulman AH. Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res 2001; 11:2041 - 2049
- San Miguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL. Nested retrotransposons in the intergenic regions of the maize genome. Science 1996; 274:765 - 768
- Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol 2006; 4:119 - 128
- Lawrence JG. Russell W, Herwald H. Horizontal and vertical gene transfer: the life history of pathogens. Concepts in Bacterial Virulence 2005; Karger Contrib Microbiol Basel 255 - 271
- Tsirigos A, Rigoutsos I. A new computational method for the detection of horizontal gene transfer events. Nucl Acids Res 2005; 33:922 - 933
- Rieseberg LH, Soltis DE. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plant 1991; 5:65 - 84
- Kishino H, Hasegawa M. Evaluation of the maximum likelyhood estimate of the evolutionary tree topologies from DNA sequence Data, and the branching order in Hominoidea. J Mol Evol 1989; 29:170 - 179
- Shimodaira H, Hasegawa M. Multiple comparisons of Log-Likelihoods with application to phylogenetic inference. Mol Biol Evol 1999; 16:1114 - 1116
- Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst Biol 2000; 49:652 - 670
- Strimmer K, Rambaut A. Inferring confidence sets of possibly misspecified gene trees. Proc R Soc Lond B 2002; 269:137 - 142
- Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 2004; 14:860 - 869
- Vitte C, Ishii T, Lamy F, Brar DS, Panaud O. Genomic paleontology provides evidence for two distinct origins of asian rice (Oryza sativa L.). Mol Genet Genom 2004; 272:504 - 511
- Vitte C, Panaud O, Quesneville H. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics 2007; 8:218
- Roulin A, Piégu B, Panaud O. Evidence of multiple horizontal transfers of the long terminal repeat retrotransposon RIRE1 within the genus Oryza. Plant J 2008; 53:950 - 959
- Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Evol 1997; 28:593 - 619
- Feschotte C. Transposable element and the evolution of regulatory networks. Nat Rev Genet 2008; 9:397 - 405
- Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin breceptor and apolipoprotein C–I genes in humans. J Biol Chem 2001; 276:1896 - 1903
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 2006; 441:87 - 90
- Shen Z, Denton M, Mutti N, Pappan K, Kanost MR, Reese JC, Reek GR. Polygalacturonase from Sitophilusoryzae: possible horizontal transfer of a pectinase gene from fungi to weevils. J Insect Sci 2003; 3:24
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A. Evidence for horizontal transmission of the P transposable element between drosophila species. Genetics 1990; 124:339 - 355
- Haring E, Hagemann S, Pinsker W. Ancient and recent invasions of drosophilids by P elements. J Mol Evol 2000; 51:577 - 586
- Clark JB, Kidwell MG. A phylogenetic perpective on P transposable element evolution in Drosophila. Proc Natl Acad Sci USA 1997; 94:11428 - 11433
- De Almeida LM, Carareto CM. Multiple events of horizontal transfer of the Minos transposable element between Drosophila species. Mol Phylogenet Evol 2005; 35:583 - 594
- King Jordan I, Matyunina LV, McDonald JF. Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc Natl Acad Sci USA 1999; 96:12621 - 12625
- Robertson HM, Lampe DJ. Recent horizontal transfer of a mariner transposable element among and between diptera neuroptera. Mol Biol Evol 1995; 12:850 - 862
- Casse N, Bui QT, Nicolas V, Renault S, Bigot Y, Laulier M. Species sympatry and horizontal transfers of Mariner transposons in marine crustacean genomes. Mol Phylogenet Evol 2006; 40:609 - 619
- Hamada M, Kido Y, Himberg M, Reist JD, Ying C, Hasegawa M, Okada N. A newly isolated family of short interspersed repetitive elements (SINEs) in coregonid fishes (whitefish) with sequences that are almost identical to those of the SmaI family of repeats: possible evidence for the horizontal transfer of SINEs. Genetics 1997; 146:355 - 367
- Volff JN, Körting C, Schartl M. Multiple lineages of the non-ltr retrotransposon Rex1 with varying success in invading fish genomes. Mol Biol Evol 2000; 17:1673 - 1684
- Laha T, Loukas A, Wattanasatitarpa S, Somprakhon J, Kewgrai N, Sithithaworn P, Kaewkes S, Mitreva M, Brindley PJ. The bandit, a new DNA transposon from a hookworm: possible horizontal genetic transfer between host and parasite. Plos Negl Trop Dis 2007; 1:35
- Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. Massive horizontal transfer of mitochondrial genes from divers land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA 2004; 101:17747 - 17752
- Mower JP, Stefanovic S, Young GJ, Palmer JD. Plant genetics: gene transfer from parasitic to host plants. Nature 2004; 432:165 - 166
- Davis CC, Wurdack KJ. Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science 2004; 305:676 - 678
- Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot 2007; 58:1 - 9
- Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, Panaud O. Doubling genome size without polyploidization: dynamics of retrotrans-position-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 2006; 16:1262 - 1269
- Kidwell MG. Lateral transfer in natural populations of eukaryotes. Annu Rev Genet 1993; 27:235 - 256
- Kashkush K, Feldman M, Levy AA. Transcriptional actiavtion of retrotransposons alters the expression of adjacent genes in wheat. Nature Genet 2003; 33:102 - 106