Abstract
A hallmark of vascular plants is the development of a complex water-conducting system, which is physically reinforced by the heterogeneous aromatic polymer lignin. Syringyl lignin, a major building block of lignin, is often thought to be uniquely characteristic of angiosperms; however, it was demonstrated over fifty years ago that that syringyl lignin is found in another group of plants, known as the lycophytes, the ancestors of which diverged from all the other vascular plant lineages 400 million years ago.1 To determine the biochemical basis for this common biosynthetic ability, we isolated and characterized cytochrome P450-dependent monooxygenases (P450s) from the lycophyte Selaginella moellendorffii and compared them to the enzyme that is required for syringyl lignin synthesis in angiosperms. Our results showed that one of these P450s encodes an enzyme that is functionally analogous to but phylogenetically independent from its angiosperm counterpart. Here, we discuss the evolution of lignin biosynthesis in vascular plants and the role of Selaginella moellendorffii in plant comparative biology and genomics.
Introduction
Land plants made their first appearance on land in the Middle Palaeozoic era, between 480 and 360 million years ago.Citation2 Fossil evidences as well as phylogenetic studies suggest that the earliest land plants were small in stature and simple in morphology, and may have resembled the bryophytes (mosses, liverworts and hornworts) living today.Citation3 It was not until the Early Devonian that the development of a complex water-conducting xylem structure, strengthened by secondary cell wall lignification, allowed tracheophytes to dominate the flora of the earth during the Carboniferous period.Citation4 The evolution of lignin biosynthesis, possibly through the elaboration of pathways originally involved in the synthesis of UV-protective pigments, was an essential step towards the successful colonization of terrestrial environments by plants.
Lignin from flowering plants is typically composed of three major building blocks, namely p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) subunits, which are derived from three phenylpropanoid alcohols, p-coumaryl, coniferyl and sinapyl alcohol, respectively, also known as monolignols. Research in phenylpropanoid metabolism, mainly in Arabidopsis and other agricultural important dicot species, has revealed that lignin biosynthesis in angiosperms requires three cytochrome P450-dependent monooxygenases (P450s).Citation5 Whereas cinnamic acid 4-hydroxylase (C4H) and p-coumaroylshikimic acid 3′-hydroxylase (C3′H) catalyze aromatic ring 4- and 3-hydroxylation reactions leading to the formation of H and G lignin, ferulic acid/coniferaldehyde/coniferyl alcohol 5-hydroxylase (F5H) diverts G lignin biosynthetic intermediates towards S lignin biosynthesis. It has frequently been considered that F5H is a recent innovation of the angiosperms because S lignin is generally absent in gymnosperms and ferns; however, S lignin has also been found in Selaginella species,Citation6–Citation10 a genus representing the lycophyte lineage that diverged from other vascular plants 400 million years ago (A and B). This observation raises the intriguing question of how plants separated by 400 million years of evolution possess a comparable lignin biosynthetic repertoire.
Independent Recruitment of a P450 to S Lignin Biosynthesis in Selaginella
The fact that Selaginella deposits S lignin suggests that its genome encodes an enzyme with activity similar to an angiosperm F5H. To determine the identity of this protein, we isolated Selaginella F5H candidate genes from a previously reported Selaginella moellendorffii cDNA library.Citation11 Each of these genes was transformed into the F5H-deficient Arabidopsis fah1–2 mutant but only one complemented all of the mutant's phenotypes,Citation12 suggesting that the encoded Selaginella P450 is a functional F5H in vivo (SmF5H). When expressed in yeast, the protein's kinetic characterization revealed that SmF5H resembles angiosperm F5Hs in that it favors coniferaldehyde and coniferyl alcohol over ferulic acid as substrates. Interestingly, SmF5H is less than 40% identical to angiosperm F5Hs; whereas, Selaginella C4H and C3′H orthologs share over 60% identity to their angiosperm counterparts. More detailed phylogenetic analysis indicated that SmF5H is not orthologous to angiosperm F5Hs, but rather is a member of a clade of eleven P450s unique to Selaginella. None of the other ten Selaginella P450s in this clade possesses F5H activity. We therefore conclude that the origins of S lignin in Selaginella and angiosperms are due to convergent evolution of P450 activities.
Has S Lignin only Evolved Twice during Plant Evolution?
Reports from the older literature suggest that S lignin might have arisen multiple times in tracheophyte lineages. Using either histochemical staining or, by today's standards, relatively crude chemical degradation methods, S lignin has been detected in fern and gymnosperm species, including cuplet fern (Dennstaedtia bipinnata), yew plum pine (Podocarpus macrophyllus), sandarac-cypress (Tetraclinis articulata), and all the three extant genera within the division Gnetophyta,Citation6,Citation13–Citation16 and we have verified the presence of S lignin in Podocarpus macrophyllus by the highly diagnostic derivatization followed by reductive cleavage method (DFRC)Citation17 (). Elucidation of the molecular mechanisms that underlie the deposition of S lignin in these plants will further advance our understanding of the evolution of both P450s and plant phenylpropanoid metabolism.
Selaginella moellendorffii as a Model for Comparative Biology
The evolution of the lycophytes, including extant species like Selaginella moellendorffii, has been independent of all the other vascular plants, collectively known as euphyllophytes, since their common ancestors diverged 400 million years ago.Citation11 Although Selaginella maintains many features that are considered to be primitive for vascular plants, including dichotomous branching pattern of both shoots and roots, microphylls without complex vein networks, and a protostele with xylem surrounded by phloemCitation18 (), Selaginella may also have evolved developmental and biochemical processes that were uniquely elaborated in the lycophyte lineage. The fact that SmF5H, as well as the other ten Selaginella P450s studied in our research, are not orthologous to any of the known P450s found in other plant lineages suggests these P450s may be representative of such novel pathways. The recent availability of the Selaginella moellendorffii genome sequence makes this species an interesting target for comparative genomics, development and biochemistry.Citation19
Using Angiosperm Model Systems to Study Selaginella Gene Function
Although the techniques for gene-knockout, knockdown and overexpression are still to be developed in Selaginella, our research and work from other labs suggests that established model systems such as Arabidopsis can be used as platforms to study Selaginella gene function in planta. For example, Harrison et al., studied the evolution of KNOX-APR interactions in determining leaf formation.Citation20 They showed that the APR homolog from Selaginella complements the Arabidopsis APR mutant as1-1, which provided in vivo evidence to support the hypothesis that Selaginella APR is functionally equivalent to its Arabidopsis counterpart.Citation20 Furthermore, two independent groups studied GID1-DELLA mediated gibberellin (GA) growth regulatory mechanisms in moss and Selaginella.Citation21,Citation22 Whereas one group showed that both Selaginella GID1 and DELLA homologs can complement the respective mutants in rice,Citation21 the other group conducted a GA inducible green fluorescent protein (GFP) fluorescence quenching assay to show that transgenic GFP-tagged Selaginella DELLA can interact with Arabidopsis GID1 in vivo.Citation22
Future Directions
Our research marks the first step towards understanding lignin biosynthesis, or phenylpropanoid metabolism in general, in Selaginella. Many other genes that encode pathway enzymes or regulatory factors remain to be characterized. Although Arabidopsis mutants for almost all the steps in the pathway are available, thus enabling studies similar to our recent work with SmF5H, development of genetic resources and techniques, such as insertion mutant lines and Agrobacterium-mediated transformation, in Selaginella is still needed. This is particularly true when comes to identifying and characterizing novel pathways that do not exist in flowering plants. From a comparative genomics point of view, the recent completion of genome sequences for Selaginella moellendorffii and the moss Physcomitrella patensCitation23 () has made it obvious that we need sequences for genomes from representative fern and gymnosperm lineages. Approaching gene functions in an evolutionary context, propelled by the development of further genomic resources, will ultimately enable us to understand how plants evolved and flourished on earth.
Figures and Tables
Figure 1 Selaginella moellendorffii represents an ancient land plant lineage. (A) A simplified cladogram illustrating the evolutionary position of Selaginella in the plant kingdom. Species with fully sequenced genomes are noted in brackets. (B) The aerial part of Selaginella moellendorffii showing dichotomous branching at the shoot. (C) Selaginella root with dichotomous branching pattern. (D) Selaginella microphylls with a single, unbranched vein emerged from the stele. (E) A cross-section of Selaginella stem showing protostelic vasculature.
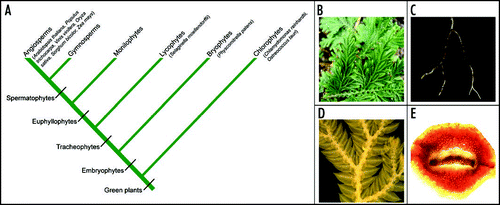
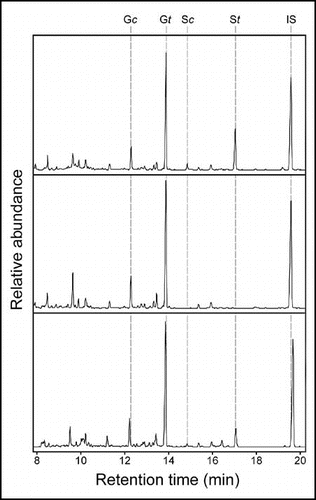
Figure 2 DFRC GC analysis of lignin monomer diversity in vascular plants. Arabidopsis Columbia wild type (top) and the fah 1–2 mutant (middle) serve as positive and negative controls for the presence of syringyl lignin. DFRC lignin analysis was performed as previously described.Citation17 DFRC analysis of a sample of Podocarpus macrophyllus (bottom) collected from the Chicago Botanic Garden reveals the presence of syringyl lignin in this gymnosperm. G/S, guaiacyl/syringyl lignin derivative; c/t: cis/trans; IS, internal standard.
Acknowledgements
This work was supported by the National Science Foundation, Grant No. IOB-0450289. This is journal paper number 2008-18355 of the Purdue University Agricultural Experiment Station.
Addendum to:
References
- Weng JK, Li X, Stout J, Chapple C. Independent origins of syringyl lignin in vascular plants. Proc Natl Acad Sci USA 2008; 105:7887 - 7892
- Stewart WN, Rothwell GW. Paleobotany and the evolution of plants 1993; New York Cambridge University Press
- Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature 1997; 389:33 - 39
- Friedman WE, Cook ME. The origin and early evolution of tracheids in vascular plants: integration of palaeobotanical and neobotanical data. Philos Trans R Soc Lond B Biol Sci 2000; 355:857 - 868
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 2003; 54:519 - 546
- Logan KJ, Thomas BA. Distribution of lignin derivatives in plants. New Phytol 1985; 99:571 - 585
- Faix O, Gyzas E, Schweers W. Comparative Investigations on Different Fern Lignins. Holzforschung 1977; 31:137 - 144
- Erickson M, Miksche GE. Characterization of Pteridophyta Lignins by Oxidative-Degradation. Holzforschung 1974; 28:157 - 159
- Towers GHN, Gibbs RD. Lignin chemistry and the taxonomy of higher plants. Nature 1953; 172:25 - 26
- White E, Towers GHN. Comparative biochemistry of lycopods. Phytochemistry 1967; 6:663 - 667
- Weng JK, Tanurdzic M, Chapple C. Functional analysis and comparative genomics of expressed sequence tags from the lycophyte Selaginella moellendorffii. BMC Genomics 2005; 6:85
- Chapple CC, Vogt T, Ellis BE, Somerville CR. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 1992; 4:1413 - 1424
- Miksche GE, Yasuda S. Gas-chromatography analysis of lignin oxidation-products 16. Lignin in leaves of several softwoods and hardwoods. Holzforschung 1977; 31:57 - 59
- Kuroda H. Comparative studies on O-methyltransferases involved in lignin biosynthesis. Wood Res 1983; 69:91 - 135
- Creighton RHJ, Gibbs RD, Hibbert H. Studies on lignin and related compounds LXXV Alkaline nitrobenzene oxidation of plant materials and application to taxonomic classification. J Am Chem Soc 1944; 66:32 - 37
- Erickson M, Miksche GE. Characterization of gymnosperm lignins by oxidative-degradation. Holzforschung 1974; 28:135 - 138
- Lu F, Ralph J. The DFRC Method for lignin analysis 2. Monomers from isolated lignins. J Agric Food Chem 1998; 46:547 - 552
- Bowman JL, Floyd SK, Sakakibara K. Green genes-comparative genomics of the green branch of life. Cell 2007; 129:229 - 234
- 2008; JGI. http://genome.jgi-psf.org/Selmo1/Selmo1.home.html
- Harrison CJ, Corley SB, Moylan EC, Alexander DL, Scotland RW, Langdale JA. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature 2005; 434:509 - 514
- Hirano K, Nakajima M, Asano K, Nishiyama T, Sakakibara H, Kojima M, Katoh E, Xiang H, Tanahashi T, Hasebe M, Banks JA, Ashikari M, Kitano H, Ueguchi-Tanaka M, Matsuoka M. The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell 2007; 19:3058 - 3079
- Yasumura Y, Crumpton-Taylor M, Fuentes S, Harberd NP. Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol 2007; 17:1225 - 1230
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin IT, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008; 319:64 - 69