Abstract
The bacterium Serratia marcescens produces a plethora of multicellular shapes of different colorations on solid substrates, allowing immediate visual detection of varieties. Such a plasticity allows studies on multicellular community scale spanning two extremes, from well-elaborated individual colonies to undifferentiated cell mass. For a single strain and medium, we obtained a range of different multicellular bodies, depending on the layout of initial plating. Four principal factors affecting the morphogenetic pathways of such bodies can be distinguished: (1) amount, density, and distribution pattern of founder cells; (2) the configuration of surrounding free medium; (3) the presence and character of other bacterial bodies sharing the same niche; and (4) self-perception, resulting in delimitation towards other bodies. The last feature results in an ability of well-formed multicellular individuals to maintain their identity upon a close mutual contact, as well as in spontaneous separation of cell masses in experimental chimeras. We propose an "embryo-like" colony model where multicellular bacterial bodies develop along genuine ontogenetic pathways inherent to the given species (clone), while external shaping forces (like nutrient gradients, pH, etc.) exert not formative, but only regulative roles in the process.
Introduction
The morphogenesis of multicellular organisms is usually exemplified by the model case of animal ontogeny, i.e., as an irreversible process resulting in an individual. Many multicellular life forms, however, might be bound by much looser morphogenetic constraints, allowing alternative developmental pathways controlled by external or internal cues as, e.g., in plants. Not only a variety of shapes can arise from a zygote or a spore; some ontogenetic strategies allow also vegetative propagation by buds, cuttings, founder cells and similar body fragments. Some life forms can thus be “parsed” (even to the level of single cells) and give rise to new shapes. We focused on such plasticity in a model prokaryotic organism—the bacterium Serratia marcescens, whose natural coloration greatly facilitates in vivo observation of developing multicellular structures.
Bacteria cover an admirable range of multicellular life forms, even within a single species or strain. They may grow as undifferentiated cell masses or suspensions; given, however, opportunities (free space, nutrients, ground texture, neighbors, etc.), they give rise to sophisticated body forms—societies created by billions of cells—often following morphogenetic pathways akin to those observed in genuine individuals.Citation1–Citation4 S. marcescens is known for its extreme variability of multicellular bodies formed under different settings.Citation5–Citation8 As nutritional conditions are a prominent factor, the variability of bodies tends to be described as an epiphenomenon of nutrient limitation and/or metabolite buildup in the medium.Citation9,Citation10 Often the morphogenesis of bacterial bodies is depicted or modeled as if molded by external forces like, e.g., the behavior of iron particles in a magnetic field, or as a crystallization-like process, with bacteria playing role of “molecules”.Citation11–Citation13 Decisive for a genuine embryonic development are, however, not the nutrients and bulk metabolites but specific signals. The participating cells are interpreting them in a context-dependent, semantic manner, which results in cell populations proceeding along inherent morphogenetic trajectories. Here we show that this indeed appears to be the case also in the development of multicellular bacterial bodies.
Results
Classification of bacterial bodies
Under standardized nutrient conditions and in the presence of glucose, Serratia marcescens CNCTS 5965 gives rise to distinctly shaped and colored colonies (see below), while only smooth red colonies were obtained on identical media without glucose. On glucose-containing media, the original bacterial population occasionally segregated clones lacking the red pigment. After several rounds of selection, we derived two stable variants of the strain, colored (F) and white (W), which no longer segregated and which have been used in all the subsequent experiments.
Surprisingly, colonies of both variants exhibited a finite growth, reaching, under optimal settings, a maximum diameter of 15 mm (the final size was somewhat smaller under suboptimal conditions, such as dense plating). Colonies and other multicellular bodies (see below) reached their final size, shape and color within 7–10 days upon plating, and survived for at least another three weeks without substantial changes ().
Upon streaking the bacteria on a dish, multicellular bodies ranging from an undifferentiated cell mass to characteristically patterned colonies could be distinguished (). For the sake of further study, we define three well-differentiated body types—colony, confluent colony and macula—that appear on such a continuum, depending on the number, density and distribution of colony-forming units (CFU) in the original plating.
A colony was provisionally defined as a multicellular body grown from a single cell. Its appearance depends on the density of plating:
Bacteria sown at hundreds CFU per dish produce many small colonies (diameter of 1–2 mm), colored or white depending on the strain used (colored colonies shown in ). As we shall discuss below, these can be viewed as incompletely developed, “dormant” colonies.
At tens CFU per dish, colonies develop to a diameter of about 10 mm, i.e., the maximum mature size at that density of plating. The colored form is characterized by a shape resembling a fountain (F), with an elevated smooth red center and an elevated red rim, separated by a rough white circle (B and C). Colonies in somewhat denser plating are smaller and produce only the red rim, but no elevated center (see ). The colonies of the white (W) strain are shaped somewhat differently ( and ).
Bodies indistinguishable from F or W colonies described above, and of the same size, arose also from multicellular inocula, such as up to 106 cells planted (dotted or dropped, as described in Methods) to a strictly limited spot. Sometimes, however, multicellular CFU of the F clone produced disheveled forms with many red protuberances (). Thus, in agreement with others (reviewed in refs. Citation14 and Citation15), a colony will be understood here not only as a clonal body grown from a single cell, but also as a morphological entity, independent from the number of founders. (This also brings a methodological advantage, since colonies can be planted to desired coordinates on the dish.) Even more importantly, mature large F and W colonies can be viewed as fully developed specimens of a lineage-specific “body plan”, while small colonies, or other forms described below, may be interpreted as instances of an incompletely accomplished development towards the prototype F or W colony pattern.
Next, we investigated initial and boundary conditions allowing development of colonies or colony-like bodies. In particular, cell density and the size and layout of the planting area (dot, drop or smear), can be understood as two dimensions of a bacterial “morphospace” that may act in mutual discordance (). Two additional characteristic shapes were observed under such “suboptimal” conditions.
Confluent colony (“concol”) develops upon spreading a bacterial suspension (approx. 15–150 cells/cm2) over an area up to a diameter of 15 mm, i.e., similar to, or slightly exceeding, the size of a mature, fully developed colony (). A concol can also arise if colonies in a sparse sowing meet prior to achieving their fully differentiated state (see and ). Three types of concols can be observed for the F strain; bodies of the W form are analogous, albeit white (see ):
At lower densities (approx. 20 cells/cm2), a small number of colonies is initiated; they soon merge, establishing a center consisting of several red spots (corresponding approximately to the number of founder cells), surrounded by an elevated common red rim reminiscent of an F colony (as in and ; compare also ).
Densities up to 150 cells/cm2 produce concols with a single granular red center surrounded by a white laced zone, which later develops into a red rim, albeit less perfect than that at lower cell densities (). Best-looking concols developed from a single spot of bacterial suspension per dish.
If the diameter of the inoculated area exceeds the 15–20 mm limit, cells no longer establish a concol, the only exception being elongated concols developing when one dimension in the spread is limited to about 7 mm (, lower left of each dish). Again, however long such a body might be, it never grows far into the unoccupied area beyond its borders.
Unlike colonies, concols do not develop in the same manner after planting multicellular inocula in a dense spacing instead of sowing a cell suspension; results of such experiments will be described in more detail below.
If the density of spreads is even higher than in concols (>150 CFU/cm2) and, at the same time, the planting area exceeds the diameter of 10–15 mm, distinguishable colonies never appear. Instead, the F bacteria develop into a macula—a red carpet without apparent inner structuring but with a well-developed rim as the last remnant of the F-type developmental pathway (, right). Best results were again obtained with a single body per dish: the development of a macula requires a certain amount of unoccupied medium in its surrounding. Smears as those in represent maculae elongated in one dimension. An analogous phenotype was obtained also for the W form.
We further focused on three factors affecting morphogenesis of bodies described: availability of unoccupied substrate, the presence of other bodies, and internal processes governing body shaping.
Behavior towards unoccupied medium
A dense suspension, spread evenly over the whole dish, results in a population of small, separated colonies ( and ). Spreading on dishes of varying diameter (9, 6, 3.4 or 1.5 cm, see ) or immediate removal of the surrounding agar field after plating the cells in a 1.5 cm disc in the center of a dish () produces the same pattern, provided the whole area is covered by the bacterial spread at a similar density. However, spreading of an inoculum of the same density in a circle of a diameter 1.5 cm in the center of a larger dish results in a concol instead of multiple isolated colonies, indicating that the free agar medium surrounding the inoculum provides some factor that facilitates colony expansion into the unoccupied space ().
An obvious explanation would be that bacteria on fully occupied plates simply cease growing due to exhaustion of the medium or buildup of metabolites. However, the same pattern of small colonies was obtained upon replacement of the removed solid medium by liquid broth, maintaining constant nutritional conditions across the whole dish (). In subsequent experiments we found that at least 1.5 cm of unoccupied medium in all directions is required for full concol development (not shown). Hence, the development of the growing concol reflects the availability of free agar in its surroundings, possibly via perceiving some physical parameter or a specific rate-limiting compound provided and/or sequestered by the agar component of the solid media, unless we assume a substantial influence of relatively minor differences in water availability in the broth-supplemented agar block. The later explanation, however, is unlikely, since even at agar concentration down to 0.5% some aspects of the F pattern can develop, albeit less efficiently than on full-strength agar (not shown).
Upon one-sided removal of the free agar in the vicinity of the inoculated central disk (), the bacterial body responds by developing into an imperfect concol (as in ) in the direction of remaining agar, whereas the cut edge remains flanked by free colonies.
Upon transfer of small (ca 1 cm2) discs cut out from a continuous carpet of mature dormant colonies (hundreds per dish; left) into an opening of identical diameter in an empty agar field (from the same batch of dishes, middle), colonies resumed their development, expanded somewhat beyond the confines of the transferred agar plug and merged into a macula. Similar discs transferred into an empty dish without agar remained unchanged ( right). Such a behavior took place even with older (24 days) cultures. (Note that a comparable number of CFU—approx. 20—sown in the middle of the dish would result in a concol, as in ; however, here we did not transfer freshly sown cells but established colonies).
Thus, bodies seemingly in a final stage of their ontogeny can resume development if challenged by fresh unoccupied medium. However, even in such a case they remain spatially restricted, suggesting the involvement of specific regulatory processes as opposed to simple nutrient limitation.
Close neighborhood effects
To examine the mutual behavior of bodies sharing the same dish, we followed the development at several plating configurations.
First, dishes were sown with a suspension of the F strain at hundreds of CFU per dish, which should produce small colonies, and an inoculum of the same strain sufficient to start a colony or a macula was planted (dropped or dotted) upon this field immediately. Whereas the colonies of the background developed normally, only an undifferentiated blotch lacking structures of a typical macula or colony developed on the planting site, while controls developed normally (). Although inhibition was observed even if the number of cells in the planting exceeded that in the field by several orders of magnitude, it was more profound on a dense field and/or upon planting a diluted suspension. As expected, dropping of inocula corresponding to a colony or macula to fields of 2–4 days old colonies led to even more dramatic impediment of development (not shown).
If such a dense bacterial field was applied only to one half of a dish, and a macula-producing inoculum was planted immediately at the interface of the occupied and free agar, a typical macula with a well-developed rim appeared in the direction of the free agar; its rim, however, disappeared at the border facing neighboring colonies (), reminiscent of the situation at the interface of free agar and empty space ().
Multiple comparably strong inocula of 102–105 cells planted close to each other developed to normal F colonies, neither inhibited nor merged into a concol (). However, colonies developing from small inocula did produce a concol (). This indicates the existence of a limit inoculum size deciding the result of a tradeoff between tendencies to merge into a concol, or to develop a full-fledged colony, at a given distance (see also the overlap of configurations in the morphospace phase diagram at ).
Long-distance signaling between bodies
F colonies incidentally growing in close vicinity develop their coloration quicker than their peers growing sparsely; from such a colored center the reddening front spreads into the surroundings (). The effect suggests that colonies can induce final phenotypes in their neighbors.
To get a closer insight, a smear of approx. 108 cells was applied to a line on agar, and, at the same time, colonies were planted 1–2 cm from that line. The smear developed into a macula that accelerated the coloring of colonies, as compared to the controls, to a degree proportional to the distance between both bodies (). A trench cut into the agar did not prevent acceleration of color development, suggesting participation of a signal traveling through the gas phase (). A macula of W cells also exerted an accelerating effect on F colony coloration ().
Surprisingly, also an F macula induced weak pink coloration in W colonies, together with disruption of colony structure (; for structure disruption see also ). Similar disruption of W colony structure was observed also in the vicinity of a W macula (not shown). Thus, an F body can induce the production of pigment in W colonies. The coloring, albeit fainter, appeared also in a configuration with the trench (not shown). Cells from the pink crescent gave rise to normal W colonies, even after five rounds of co-cultivation with an F macula as described above, indicating a transient, regulatory nature of the observed changes.
Chimeric bodies
Upon collision of growing F and W colonies, the two bodies do not merge into a concol with a common rim but maintain a clear-cut boundary instead. As in the case described above, the W colony develops a pink coloration adjacent to the F neighbor (). While this phenomenon might be analogous to the above-described collision of large colonies that also do not merge (), it prompted us to investigate the behavior of bodies originating from mixed suspensions.
When dense suspensions containing a mixture of cells of F and W lineages were dropped in a manner that would produce colonies under monoclonal conditions, colony-sized chimerical bodies (i.e., about 10–12 mm in diameter) lacking the typical F morphology developed, with a red center and white periphery. The proportion of colors roughly corresponded to the ratio of F and W cells in the original suspension ( and middle). Material dotted directly from the red or white part of a chimera, or sown from suspensions prepared from local samples, maintained the characteristics of the place it was taken from, suggesting segregation of both constituting clones into delimited territories, while a suspension from the whole chimera gave rise to a mixture of F and W colonies (C and D). Chimerism was further confirmed by pattern reconstitution in chimeras disrupted in situ using a sterile glass rod at 1, 2 and 4 days after planting (i.e., prior to development of coloration, which was only starting to appear in the oldest bodies). 2–4 days after such homogenization the body re-established its red center and white margin (). The younger the chimera at the time of disruption, the more complete was the pattern regeneration; in 4 days-old chimeras, the regeneration capacity is nearly lost (see bottom row of ).
When bacteria from white or red parts of such regenerated bodies were replanted, they produced colonies whose character corresponded to their kin in the region the dot was taken from, indicating that reconstitution involves cell migration rather than phenotype switching in individual cells. Material from red parts of incompletely reconstituted older (4 days) chimeras produced mixed bodies, consistent with imperfect redistribution of both cell types, possibly due to increasing rigidity of older chimeras, which impedes their complete homogenization.
Unlike chimeras, F colonies did not regenerate their original pattern after homogenization at 2 to 3 days, producing only variegated bodies (). Obviously, cells of the same clonal origin do not perceive their sisters as “strangers”, regardless of their (color) differentiation status, and, thus, they do not migrate to segregate the two colored variants.
Discussion
Prior to the advent of the DNA era, bacterial colonies were mostly viewed as epiphenomena or diagnostic tools revealing “true” performances taking place at cellular level, such as pathogenicity, biochemical processes, mutations or even life cycles (reviewed in ref. Citation16). For example, Mary I. Bunting in a series of papers on colony plasticity (so called “dissociation”) in SerratiaCitation5–Citation8 seeks after trends going on not in colonies proper, but in suspensions that gave birth to such colonies. (For a review written as an epilogue of the whole period (see ref. Citation17).
Post-war bacterial genetics brought a somewhat higher status to bacterial bodies, especially after the discovery of true life cycles (e.g., Bacillus, Caulobacter), often encompassing multicellular stages with multiple cell types (e.g., Myxobacteria, Streptomyces, Cyanobacteria). Finally, multi-species consortia such as mats, plaques, stromatolites, etc., joined the company. A colony is no longer perceived as a heap of cells, which stick together only because they cannot disperse quickly enough. Finally, in the 1980s ShapiroCitation18–Citation20 introduced multicellularity of colonies to common awareness.
The structure of multicellular bacterial bodies might simply reflect environmental differences—often transient—caused by metabolism of growing bacterial masses, such as nutrient limitation or accumulation of various metabolites in the medium, which has been the standard explanation in earlier works.Citation21,Citation22 Shapiro,Citation14,Citation19,Citation20 on the other hand, takes the radial symmetry as one of the arguments suggesting developmental coordination of processes within the bacterial body (such as synchronous switching of transposons), i.e., active participation of cell assemblies in true morphogenetic processes. A plethora of colony shapes can also be acquired by treatment as simple as changing the density of agar support.Citation11 The recent studies of E. Ben Jacob and others go even further, proposing genuine semantical communication between bacteria comprising the colony.Citation23
Here we suggest viewing the development of multicellular bacterial bodies as a genuine counterpart of animal, plant or fungal ontogeny, i.e., production of multicellular bodies through intricate morphogenetic processes resulting from cell cooperation and communication that channel the development along species- or lineage-specific pathway (a “body plan”). We observed that bacteria can (i) produce multicellular bodies of limited size that adjust their development according to such parameters as layout of the inoculum, availability of unoccupied solid medium, or the presence of other bodies in the vicinity and (ii) distinguish between cells of identical and non-identical clonal origin upon co-cultivation. This suggests involvement of specific signals that are perceived in a context-dependent manner, similar to morphogenetic signals of multicellular eukaryotes.
“Embryology” of colonies
Bacteria in general (and our model Serratia marcescens in particular) are known to be inherently phenotypically plastic—minute variation in the environment changes the overall appearance of a multicellular body. Indeed, different colony phenotypes can be obtained in S. marcescens upon minor variation of media composition, including, e.g., variation in agar density—reviewed in refs. Citation24 and Citation25; and our unpublished data. However, here we focused on factors affecting the development of multicellular bodies under constant nutrient conditions on rich media, such as plating geometry and mutual interaction of bacterial bodies over short or long distance.
To our surprise, we found that our strain exhibits restricted growth, producing colonies whose size usually does not exceed the limit of 10–12 mm in diameter, while most strains described in the literature display unlimited growth of colonies, resulting in very big bodies.Citation26,Citation27 Also some other isolates of S. marcescens exhibit unlimited growth under the same conditions in our laboratory (not shown), indicating that the observed limitation is not due to exhaustion of the medium. It can be speculated whether such a limited growth does not result from domestication of a sort; domestication effects of laboratory life were described in bacteriaCitation15 as well as in yeast colonies.Citation28 From our point of view, finite growth further justifies the embryological analogy, with fully developed mature colonies as final, “accomplished” stages of a genuine morphogenetic pathway, consummated and modified according to the parameters of the environment (a “morphosphace” where geometry of the plating, chemical composition and its time course represent just some of multiple variables). Alternative developmental outcomes may then be viewed as different stages and/or endpoints of an endeavor towards such a final stage. In particular, three such alternatives deserve closer attention.
The first variant outcome is represented by small colonies that develop at densely populated plates, reaching their final appearance rapidly, which suggests an early quorum-based decision. Such colonies might be compared to “dormant” embryos, as frequently encountered in plants or some animals. Dormancy, however, can be overcome by discontinuing the supply of the quorum signal, e.g., by transfer to a fresh medium. At even higher plating densities, distinct colonies can no longer be established, and an unstructured lawn develops instead (not shown).
The quorum signal is unlikely to be constituted by a simple parameter such as concentration of nutrients in the medium or a signal molecule in the gas phase above it, since colonies apparently perceive directional information about the availability of unoccupied substrate, even if they do not attempt to expand onto it to a larger extent (see below). Studies on quorum signaling in Serratia revealed a plethora of specific chemical signals regulating the bacterial phenotype, such as, e.g., various acyl homoserine lactones. However, these phenomena were studied on bacteria grown in suspensions, i.e., the traits followed were biochemical.Citation29,Citation30 (Similarly, when Engelbert-Kulka et al.Citation31,Citation32 speak about the role of programmed cell death in multicellular bacterial bodies, they mean bacterial suspensions).
The decision between the pathway towards fully developed colonies and the “dormant” ones does not depend on the overall number of cells on a dish but on the amount of colony seeds (colony-forming units, CFU) that may be either unicellular or multicellular. Surprisingly, even large multicellular CFU containing millions of cells “respect” the quorum established by multiple unicellular initiating centers that make up only a negligible fraction of total cells present on the dish, thus further supporting the notion of specific signaling.
It is well documented (reviewed in refs. Citation14 and Citation33) that fully developed colonies can arise not only clonally from single cells, but also from a dense suspension, planted to a small area by dotting or dropping. Such a behavior reminds of “regulatory embryos” (like, e.g., those of mammals), which can start not only from a zygote but also from greater embryonic-cell masses or even chimaeras of different genetic backgrounds, or of regeneration of an anatomically complete plant from adventitious roots or shoots developing on a callus. However, establishing the colony body plan from such cell masses may be error-prone, as documented by occasional aberrant colonies. It also apparently takes some time before a colony from a multicellular inoculum acquires the ability to process environmental information, e.g., to monitor the presence of close neighbors. Thus, fully developed colonies form even in situations that soon lead to collision of growing bodies originated from closely planted large inocula.
Plating of a dense suspension to an area greater than the maximum colony size leads to another variant developmental outcome—confluent colonies—that may be understood as a tradeoff between several suboptimal options. The development cannot get canalized to dormant small colonies, because of lack of the necessary quorum (too few CFU) and a vast area of free substrate. For the same reason, the population cannot grow wild towards an unstructured cell mass. At the beginning, the CFU interpret their situation as favorable for establishing fully developed colonies (low CFU density plus free substrate). In the prime of the morphogenetic process, however, their growth becomes intersected with that of their peers, but at this stage it is already too late to withdraw and channel the development toward the—perhaps more appropriate—alternative pathway towards small dormant colonies; moreover, free surrounding space would not allow for such a trajectory anyway. The solution is an attempt to share the formative activities of several nuclei, resulting in a modification of the colony pattern. It is worth noting that the stronger the founders, and the closer they are planted, the better is the symmetry preserved. We interpret these outcomes as cases of teleonomic morphogenesis whereof a single fully developed colony is a fully accomplished end-point.
Finally, planting of a dense suspension to an area exceeding the size of a developed colony results, over a range of cell suspension densities, in a macula—a body whose single hint to the mature pattern may be its elevated rim.
We interpret all the developmental outcomes discussed above as products of mutual cooperation of cells within the body, i.e., not only results of passive patterning due to some external forming forces (gradients of physical parameters and/or nutrients, limitation for space, or phenomena that could be described using metaphors like crystallization).
Effects of inoculum layout
Our observations suggest that colonies adjust their development according to the parameters of surrounding space. A decisive physical or chemical cue may be provided by free, unoccupied agar medium. Moreover, this cue is likely to be specific, since if such information were carried by absolute concentration of nutrients (or by dilution of an inhibitory signal molecule), it would be provided also by liquid broth replacing the agar, or even by agar removal in case of an inhibitor. However, it has to be noted that even simple small molecules including bulk metabolites may play the part of a specific signal if they are “understood” as such (e.g., ethylene as a plant signaling molecule, or ammonia governing the morphogenesis of yeast coloniesCitation9).
Multicellular communities obviously cannot arise in liquid media, as these do not provide structured environment with gradients of nutrients, information, and—later—support for the resulting bodies. Only a semi-solid support (e.g., agar) allows establishing (often transient) spatial structures that may mold more permanent multicellular bacterial bodies. A single colony on the dish may impose to its surroundings a steep gradient (of any compound), whose slope conveys information on extent of free space available to establish a big and well-structured colony. On the contrary, dense sowings may hinder formation of such steep gradients, resulting in the choice of the pathway towards small, unstructured “dormant” bodies. Such explanations, which may deserve a more formal treatment in terms of Turing-style reaction-diffusion models, may be adopted also for cases of more complex plating layouts such as e.g., sowings to an already occupied background.
Besides self-structuring, the growing bodies also influence the development of their neighbors, at least partly via signals propagated by the gas phase, similar to the case of yeast colonies.Citation9 Searches for the “material basis” of structured gradients in the environment identified multiple candidate factors in yeasts;Citation34 a similar outcome can be predicted also for bacteria, but remains to be proved experimentally.
A natural white variety derived from our pigmented Serratia strain allowed a deeper insight into the cell and body communication phenomena. Reddening of white colonies in the vicinity of a red streak suggests that the white clone retained its capacity to synthesize the pigment, but lost the ability to generate a releasing signal, thus developing the color only in the presence of signal(s) coming from outside. This feature is stable, as no signs of genetic assimilationCitation35,Citation36 were observed. Like the famous triton axolotl (Ambystoma), white colonies remain as if locked at a “larval” stage, never proceeding to the fully developed coloring. (An alternative explanation—transfer of the dye from neighboring red cells—is extremely unlikely since the color-inducing signal is airborne).
Genetic mutations, in particular those caused by transposable elements, or stable regulatory states, can also cause morphological changes in microbial colonies.Citation14,Citation15,Citation19,Citation33,Citation37 A population of E. coli was found to generate three metabolically distinguishable forms, which persist, in constant proportions, through hundreds of generations, and which specialize for distinct metabolic tasks while “crossfeeding” the rest of the population.Citation38 Older authors also described such “differentiation-like” phenomena, showing that a suspension in its “optimal” phase produces colonies with greatest heterogeneity. Even for bacteria, true ontogeny where the developing body decides for alternative developmental trajectories towards different endpoints has been proposed in older works,Citation16,Citation39,Citation40 opening the space for recognition and analysis of phenomena such as developmental plasticity or reaction norm of the strain. Indeed, we are inclined towards such an explanation of our results.
According to M. Barbieri,Citation41 ontogeny represents a build-up of new information by re-iterating inputs from both genome and environment in an “organic memory”, which is virtually “empty” at the beginning of development. Elaborated embryonic and organogenic processes in multicellular eukaryotes are highly dependent on genetic regulatory networks (reviewed in ref. Citation42). Since the complexity of such possible networks in bacteria is far lower than that found in eukaryotes, it can be speculated that in bacteria the memory is more emancipated (less “hard-wired” into the genome).
Self/non-self recognition
We used the availability of two stable colored lines of our strain to investigate the behavior of bacterial bodies towards populations of the same species but different clonal origin. To our surprise, mixed suspensions attempted, more or less successfully, to segregate the two constituent clones into separate populations. Regeneration experiments confirmed that segregation is due to cell migration and not phenotypic switching or selective cell death. The ability of bacteria to travel long distance just to be able to reestablish zones of their own kin is an unexpected finding, and resembles a similar incompatibility observed in embryonic cells in cultures (a mixture of, e.g., ectoderm and mesoderm cells segregates in a similar manner). Moreover, it suggests that the ability to recognize “self” (i.e., cells of the same clone) from non-self (cells of related but distinct clonal origins), that has been even suggested as one of the criteria for assessing intelligence of a biological system,Citation43 may be characteristic not only for multicellular animals or plants, but also for bacteria—and, indeed, possibly for all forms of life. The capacity for intraspecific self/non-self recognition may present one of the ancestral and fundamental features of living beings.
We believe that our observations document the existence of surprisingly sophisticated communicative potential within and between bacterial colonies that presents a challenge both for experimental and theoretical biologists. One possible pathway of future investigation obviously leads towards identification of the signaling compounds and molecular mechanisms involved. Theoretical (mathematical) models of colony development also should be able to incorporate or predict the observed colony/space interaction phenomena.
Materials and Methods
Strain and media
The strain S. marcescens CNCTS 5965 was obtained from the Czech National Institute of Health, and maintained in nutrient broth No. 2 (Imuna Pharm, ZB No. 2, containing 5 g beef extract, 5 g peptone, 6.25 g nutrient base No. 1, 6.25 nutrient base No. 2, 2.5 g NaCl per litre) with 15% glycerol at -80°C. Upon reviving from the frozen stock, the bacteria were colony-purified twice and their colony phenotype was verified prior to subsequent experiments.
Bacteria were grown under standard conditions comprising 9 cm dishes with a 4% nutrient broth with nutrient agar No. 2 (Imuna Pharm ZA No. 2, i.e., ZB No. 2 with 1.5% agar), complemented with 0.5% glucose unless stated otherwise. For liquid cultures, nutrient broth No. 2 was also used.
Inoculation procedures
All manipulations were performed in a laminar flow box, at room temperature. Cell suspensions for plating were prepared in PBS (22 mM KH2PO4, 50 mM Na2HPO4, 8.6 mM NaCl, 18.7 mM NH4Cl; pH 7.4) from agar-grown mature but less than 3 weeks old bacterial bodies. Cell density of suspensions was adjusted according to optical density at 450 nm, calibrated by plating serial dilutions.
Methods of application to the agar surface were as follows:
Sowing (standard procedure, used unless specified otherwise): 100 µl of an adequately diluted bacterial suspension was spread over the whole surface of the 9 cm dish. Proportionally smaller volumes were applied if sowing to a smaller area (e.g., 40 µl to a 6 cm dish).
Dropping: a dense suspension was applied as a small drop (ca 1 µl) within a diameter of about 2 mm.
Dotting: Small amount of material was picked from a bacterial body using a sterile needle and transferred to a fresh surface.
Smear: 30 µl of bacterial suspension (approx. 108 cells) was applied to a line of approx. 5 cm.
In all cases, the suspensions soaked into agar within 2–3 min. Dishes were kept at 27°C for 2 days, and maintained at room temperature (ca 22°C) in closed but not sealed boxes containing a damp sponge to maintain humidity, wrapped in cloth to prevent insect infestation.
Documentation
Cultures were photographed using an Olympus digital camera under ambient light (except that has been taken under artificial illumination with the aid of an Olympus dissecting microscope); black background was used for visualizing white colonies in some cases. Colony size measurements have been performed on the digital photographs, either manually or with the aid of the ImageJ 1.32i software (http://rsb.info.nih.gov/ij/).
All experiments were done repeatedly; figures shown were selected from an extensive collection of primary photos. Corel Photo Paint was used to assemble the plates but no image enhancement was performed.
Figures and Tables
Figure 1 Colonies of the F clone exhibit limited growth. Colonies were started by sowing (open symbols) or dropping (closed symbols; see Methods) at time 0; average diameter from 6 colonies ± standard deviation is shown for each time point.
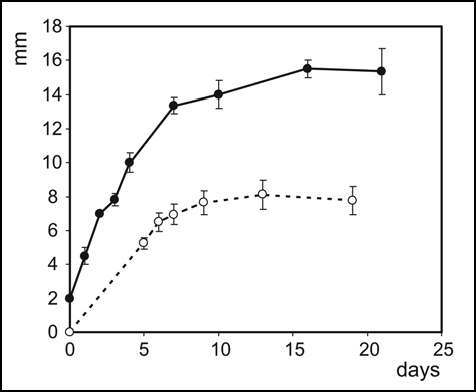
Figure 2 Continuum of multicellular bodies, from an undifferentiated cell mass to characteristically patterned colonies, develops upon streaking the bacteria on solid media. (A) Colored (F) strain; (B) white (W) strain.
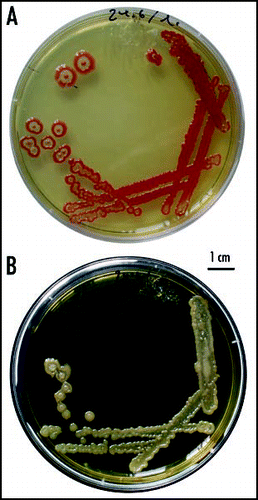
Figure 3 Types of colonies. (A) A dense spread gives rise to small colonies (the F strain, 14 days); (B) a low-density spread (about 30 CFU/dish) leads to the fountain (F) pattern (12 days); (C) a typical F colony at higher magnification with a profile of its cross-section (not to scale; 12 days); (D) a low-density spread of the W strain (6 days); (E) colonies originated by planting dense inocula: top—F colony; middle—W colony; bottom—disheveled morphology obtained sometimes for the F strain (6–9 days).
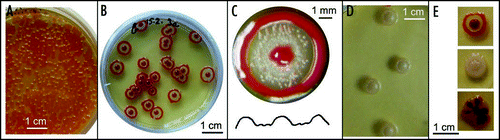
Figure 4 A “phase diagram” of the colony morphospace. Depending on the number of cells plated and the diameter of the (circular) area sown, bacteria develop into well-developed colonies (Col), confluent colonies (Con), maculae (Mac) or small, “dormant”, colonies (Dorm).
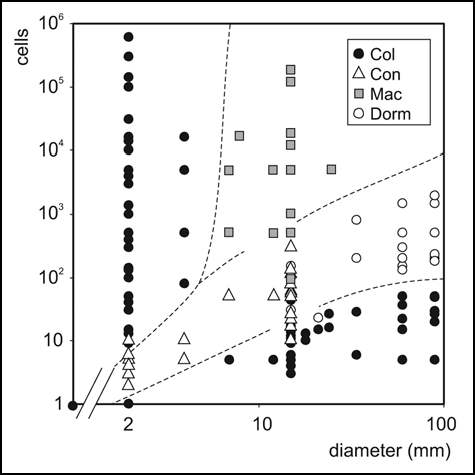
Figure 5 Bodies originating from multicellular inocula. (A) Development of a confluent colony (concol) of the F strain. Three subsequent stages at day 4, 11 and 14 after plating. (B) Spreading of a dense inoculum produces maculae. Development of 3 identical cell suspensions (about 106) spread to an area of a diameter 2, 7 and 20 mm, respectively, at 9 days after plating. Whereas two bodies at the left and middle grew while differentiating and exhibited (to a varying extent) aspects of the F colony phenotype, the body to the right did not grow beyond the original area of spread, and produced a macula.
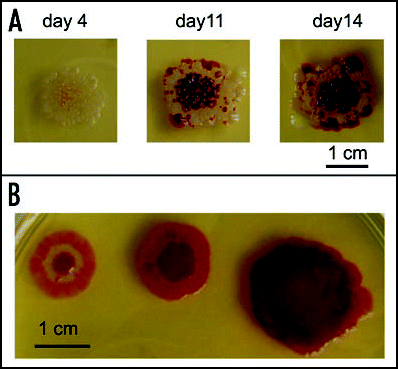
Figure 6 Effects of plating geometry. (A) Growth of dense colonies (about 300 CFU/dish) sown evenly over the whole surface of agar; (B) removal of surrounding agar leads to development of single colonies (wet tampons maintaining humidity can be seen in the dish); (C) a concol grown from a suspension of the same plating density, surrounded by free agar; (D) nutrient broth is not a sufficient substitution for the agar removed; (E) a combination of a concol and colonies developing at the interface of free space and agar. Cultures are at least 20 days old, but the same phenomena are well-developed already after 10–14 days.
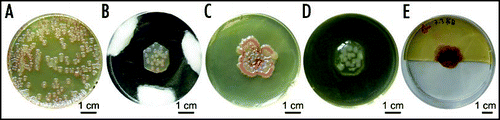
Figure 7 Transfer of colonies. Circular plugs were removed from a field of 4-day colonies (left) and transferred into a hole in a fresh agar dish (middle), or to a damp chamber (right). Fresh agar induced colonies to merge into a macula within 8 days (all dishes photographed at the same time).
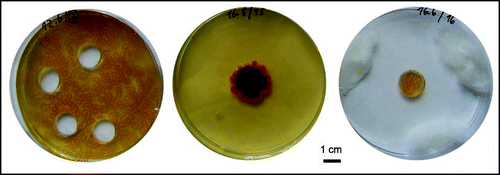
Figure 8 Development of bodies on dense bacterial fields. (A and B) Maculae from about 1500 cells, (C and D) those from about 150 cells, sown on (A and C) free agar and (B and D) a background of about 200 CFU per dish. (E and F) A similar layout with inocula normally giving rise to F colonies (3000 cells above, 300 below, respectively); (A–F) 36 days, but identical results after 14 days. (G) Development of a macula at the interface of small-colony field and free agar (6 days). (H) Three high-density drops (100–150 cells) in close vicinity develop to delimited F phenotypes, without merging into a concol; the same result was obtained with larger inocula up to 105 cells, while (I) drops containing only 10–15 cells in the same configuration give rise to a typical concol (H and I, 15 days).
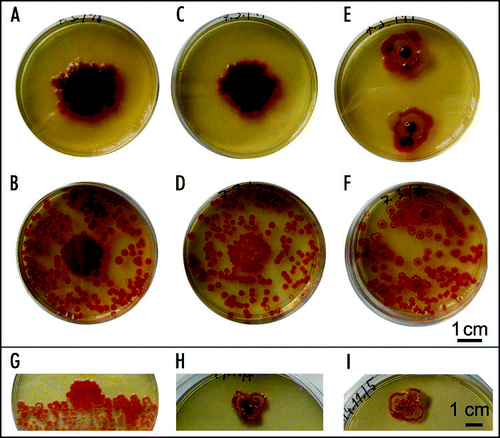
Figure 9 Spread of colony coloring from “reddening centers”. The same dish is shown at three time points.
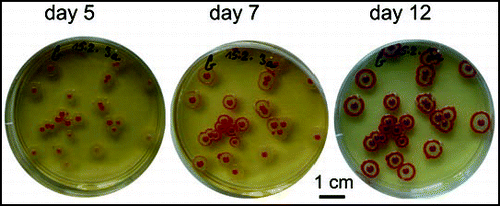
Figure 10 Effect of a neighboring macula on the development of colonies. (A) F colonies planted 1 and 2 cm, respectively, from an F macula; control (colonies only) to the right; (B) macula accelerates color development across a trench in agar; (C) a W macula accelerates ripening of F colonies; (D) W colonies adjacent to an F macula develop a faint coloring. All preparations are approx. 4 days old; snapshots were taken at the time of maximum difference between control and colonies close to a macula. After prolonged incubation all F colonies developed the standard pattern while W colonies did not acquire any additional structures.
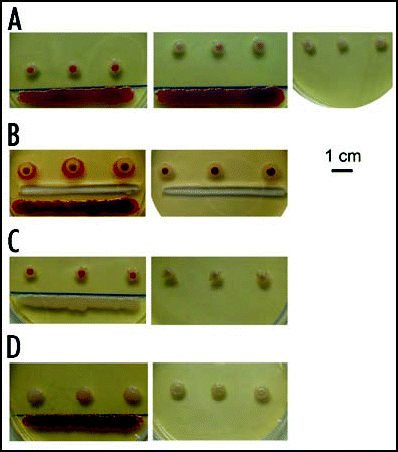
Figure 11 Encounter of colonies and development of WF chimeras. (A) W and F colonies maintain their identity upon encounter. (B) Chimeric bodies grown from mixed suspensions of W and F cells in the indicated proportions; (C) colonies dotted from the red center (left) and white rim (right) of a chimeric body; (D) colonies sown from a suspension prepared from the red center, white rim, and the whole chimera, respectively. All colonies about 10 days old.
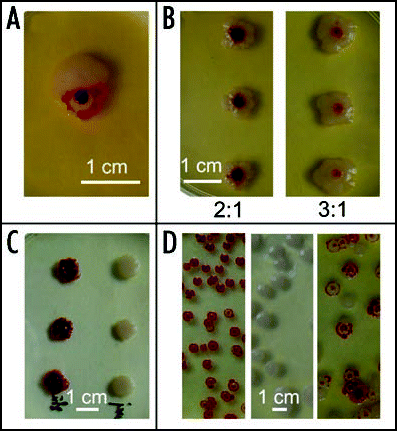
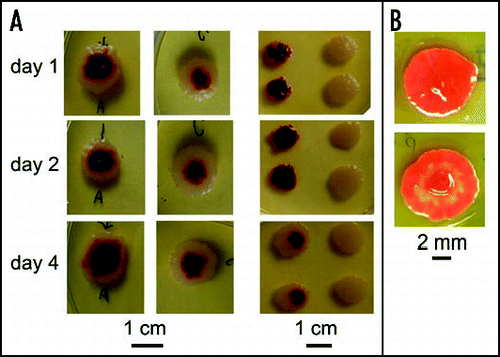
Figure 12 In situ homogenization and regeneration of chimeric and monoclonal bodies. (A) Left: WF (1:1) chimeras disrupted after 1, 2 and 4 days, respectively, and regenerated; middle: undisturbed controls; right: progeny of the reconstituted bodies (samples taken from white margins and/or red centers). For progeny of corresponding controls, see . (B) A colony that has been disturbed at 3 days photographed 6 days later (top), compared to an undisturbed control (bottom).
Abbreviations
| CFU | = | colony-forming unit |
| F | = | “fountain” (colored) colony strain |
| W | = | white colony strain |
Acknowledgements
This work was supported by the projects GACR 408/08/0796 and MSM 0021620845 to Anton Markoš, Anna Blahůšková and Zdeněk Neubauer, and MSM 0021620858 to Fatima Cvrčková. We thank Alexander Nemec for providing the bacterial strain and Vladimír Vondrejs for critical reading of the manuscript. The authors declare that they have no financial interests in the outcome of this work, nor they are aware of any conflict of interests.
References
- Ben-Jacob E, Levine H. Self-engineering capabilities of bacteria. J R Soc Interface 2005; 3:197 - 214
- Cohen I, Ron IG, Ben-Jacob E. From branching to nebula patterning during colonial development of the Paenibacillus alvei bacteria. Physica A 2000; 286:321 - 336
- Di Franco C, Beccari E, Santini T, Pisaneschi G, Tecce G. Colony shape as a genetic trait in the pattern-forming Bacillus mycoides. BMC Microbiology 2002; 2:33
- Pipe LZ, Grimson MJ. Spatial-temporal modelling of bacterial colony growth on solid media. Molecular BioSystems 2008; 4:192 - 198
- Bunting MI. A description of some color variants produced by Serratia marcescens, strain 274. J Bacteriol 1940; 40:57 - 68
- Bunting MI. The production of stable populations of color variants of Serratia marcescens #274 in rapidly growing cultures. J Bacteriol 1940; 40:69 - 81
- Bunting MI, Ingraham LJ. The distribution of color variants in ageing broth cultures of Serratia marcescens #274. J Bacteriol 1942; 43:585 - 591
- Bunting MI. Factors affecting the distribution of color variants in ageing broth cultures of Serratia marcescens #274. J Bacteriol 1942; 43:593 - 606
- Palková Z, Devaux F, Řičicová M, Mináriková L, Le Crom S, Jacq C. Ammonia pulses and metabolic oscillations guide yeast colony development. Mol Biol Cell 2002; 13:3901 - 3914
- Palková Z, Váchová L. Ammonia signaling in yeast colony formation. Intl Rev Cytology 2003; 225:229 - 272
- Matsushita M. Dworkin J, Shapiro JA. Formation of colony patterns by a bacterial cell population. Bacteria as multicellular organisms 1997; Oxford, UK Oxford University Press 366 - 393
- Li B, Wang J, Wang B, Liu W, Wu Z. Computer simulations of bacterial-colony formation. Europhys Lett 1995; 30:239 - 243
- Wakano JY, et al. Self-organized pattern formation of a bacteria colony modeled by a reaction diffusion system and nucleation theory. Phys Rev Lett 2003; 25:258102 - 258104
- Shapiro JA. Dworkin M, Shapiro JA. Multicellularity: The rule, not the exception. Lessons from E. coli colonies. Bacteria as Multicellular Organisms 1997; Oxford UP 14 - 49
- Aguilar C, Vlamakis H, Losick R, Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol 2007; 10:638 - 643
- Hadley P. Further advances in the study of microbic dissociation. J Infect Dis 1937; 60:129 - 192
- Braun W. Dissociation in Brucella abortus: a demonstration of the role of inherent and environmental factors in bacterial variation. J Bacteriol 1946; 52:243 - 249
- Shapiro JA. Bacteria as multicellular organisms. Sci Am 1988; 256:62 - 69
- Shapiro JA. Thinking about bacterial populations as multicellular organism. Ann Rev Microbiol 1998; 52:81 - 104
- Shapiro JA. Bacteria are small but not stupid: cognition, natural genetic engineering and socio-bacteriology. Stud Hist Phil Biol Biomed Sci 2007; 38:807 - 819
- Ben-Jacob E, Shmueli H, Shochet O, Tenenbaum A. Adaptive self-organization during growth of bacterial colonies. Physica 1992; 187:378 - 424
- Ben-Jacob E, Schochet O, Tenenbaum A, Cohen I, Czirok A, Vicsek T. Generic modeling of cooperative growth patterns in bacterial colonies. Nature 1994; 368:46 - 49
- Ben-Jacob E, Becker I, Shapira Y, Levine H. Bacterial linguistic communication and social intelligence. Trends Microbiol 2004; 12:366 - 372
- Lacasta AM, Cantalapiedra IR, Auguet CE, Penaranda A, Ramirez-Piscina L. Modeling of spatiotemporal patterns in bacterial colonies. Phys Rev E 1999; 59:7036 - 7041
- Giri AV, Anandkumar N, Muthukumaran G, Penathur G. A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiology 2004; 4:11
- Wimpenny JW. The growth and form of bacterial colonies. J Gen Microbiol 1979; 114:483 - 486
- Wimpenny J. Guerrero R, Pedros-Alio C. Spatial gradients in microbial ecosystems. Trends in Microbial Ecology. Spanish Soc Microbiol 1993; 135 - 140
- Kuthan M, Devaux F, Janderová B, Slaninová I, Jacq C, Palková Z. Domestication of wild Saccharomyces cerevisiae is accompanied by changes in gene expression and colony morphology. Mol Microbiol 2003; 47:745 - 754
- Labbate M, Queck SY, Koh SK, Rice SA, Givskov M, Kjelleberg S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J Bacteriol 2004; 186:692 - 698
- Van Houdt R, Givskov M, Michiels CV. Quorum sensing in Serratia. FEMS Microbiol Rev 2007; 31:407 - 424
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior of bacteria. PloS Genetics 2006; 10:1518 - 1526
- Kolodkin-Gal I, et al. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 2007; 318:652 - 655
- Shapiro JA. A role for the Clp protease in activating Mu-mediated DNA rearrangements. J Bacteriol 1993; 175:2625 - 2631
- Zikánová B, Kuthan M, Řičicová M, Forstová J, Palková Z. Amino acids control ammonia pulses in yeast colonies. Biochem Biophys Res Commun 2002; 294:962 - 967
- Waddington CH. Genetic assimilation. Adv Genet 1961; 10:257 - 290
- Waddington CH. The evolution of adaptations. Endeavour 1953; 12:134 - 139
- Siguier P, Filée J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol 2006; 9:526 - 531
- Rosenzweig RF, Adams J. Microbial adaptation to a changeable environment: Cell-cell interactions mediate physiological and genetic differentiation. BioEssays 1994; 16:715 - 717
- Braun W. Bacterial dissociation. Bacteriol Rev 1947; 11:14 - 75
- Mellon RR. The polyphasic potencies of the bacterial cell; general biologic and chemotherapeutic significance. J Bacteriol 1942; 46
- Barbieri M. The organic codes. The birth of semantic biology 2003; Cambridge, UK Cambridge University Press
- Davidson EH. Genomic regulatory systems. Dev Evol AP 2001;
- Trewavas A. Plant intelligence. Naturwissenschaften 2005; 92:401 - 413