Abstract
Pseudomonas aeruginosa can grow either as planktonic- or biofilm-form in response to environmental changes. Recent studies show that switching from biofilm to planktonic lifestyle requires rhamnolipids. Here we report the identification of a novel two-component system BqsS-BqsR that regulates biofilm decay in P. aeruginosa. BqsS is a multidomain sensor kinase and BqsR is an OmpR-like response regulator. Deletion of either bqsS or bqsR in P. aeruginosa mPAO1 resulted in a significant increase in biofilm formation. Time course analysis showed that the bqsS-bqsR mutants were defective in biofilm dispersal and in rhamnolipid production. Mutation of the BqsS-BqsR two-component system did not affect the biosynthesis of long chain quorum sensing (QS) signal N-3-oxo-dodecanoyl-homoserine lactone (3OC12HSL) but resulted in reduced production of the short chain QS signal N-butyryl-L-homoserine lactone (C4HSL) and the Pseudomonas quinolone signal (PQS). Exogenous addition of either C4HSL, PQS or rhamnolipids to the bqsS mutant reduced the biofilm formation to the wild type level. Evidence suggests that the BqsS-BqsR two-component system might promote conversion of anthranilate to PQS. Taken together, these results establish BqsS-BqsR as a novel two-component system that regulates biofilm decay in P. aeruginosa by modulating biosynthesis of QS signals and rhamnolipids.
Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium that can survive and proliferate in diverse environments. The pathogen can cause severe infections in immunocompromised individuals suffering from cystic fibrosis, burns and wounds. Because of its notorious resistance to various common antibiotics, P. aeruginosa infection has become a severe concern in intensive care units.Citation1,Citation2P. aeruginosa produces diverse virulence factors, such as exotoxin, exoenzymes, pyocyanin, rhamnolipids and lipopolysaccharide, which are the key virulence determinants in acute infection. In chronic infection, P. aeruginosa seems to adopt a surface-attached life style—biofilms. Formation of biofilms, in many cases, drastically increases the bacterial resistance to antibiotics treatment and host immune responses.Citation1 As such, understanding the genetic basis and molecular regulatory mechanisms of P. aeruginosa involved in biofilm formation and development would be essential for control and prevention of biofilm formation, and for treatment of P. aeruginosa infections.
In response to environmental changes, P. aeruginosa can switch between planktonic growth and biofilm lifestyle. Biofilm formation is a dynamic process that involves several stages, including initial attachment, microcolony formation, biofilm maturation and ultimately biofilm dispersion.Citation3–Citation6 Dispersion is an important, but less understood stage of biofilm development, in which a subpopulation of biofilm cells detach and swim away, reverting to a planktonic lifestyle. Detachment/dispersion of cells from biofilms is essential for the maintenance and continuation of biofilms.Citation7 In addition, dispersion also plays an important role in pathogenesis as the process creates mobile bacteria (single cells or aggregates) that can cause infection and promote dissemination from an initial infection point to other sites.Citation8 Dispersion is a complicated process that involves multiple steps, including degradation of biofilm matrix, activation of motility and physiological changes, which prepare cells for the conditions outside biofilms. Several environmental and biochemical factors have been demonstrated to influence biofilm dispersion, such as availability of nutrients that promotes bacterial cells to move out of biofilms,Citation9–Citation12 extracellular hydrolytic enzymes that degrade biofilm matrix leading to release of bacterial cells,Citation13,Citation14 and production of rhamnolipids, which serve as biosurfactants to interfere with cell-cell and cell-substratum interactions by disrupting reversible adhesion, irreversible attachment and influencing structural biofilm development.Citation7,Citation8,Citation15
Quorum sensing (QS) has been shown to play a role in P. aeruginosa biofilm formation under certain conditions. Mutation of the lasI gene that encodes the biosynthesis of the long-chain QS signal N-3-oxo-dodecanoyl-homoserine lactone (3OC12HSL), leads to formation of flat, undifferentiated biofilms.Citation16 While in another experiment, the lasI mutant biofilms were found indistinguishable from the wild-type biofilms at all time points with respect to both average thickness and roughness.Citation17 In addition, the short chain QS signal N-butyryl-L-homoserine lactone (C4HSL) has been proposed as a mediator of biofilm dispersion for its role in regulation of rhamnolipids production.Citation7,Citation18 In this study, we demonstrate that a novel two-component regulatory system BqsS-BqsR plays a critical role in regulation of biofilm decay in P. aeruginosa. We showed that the null mutants of the two-component system were defective in biofilm decay and in rhamnolipid production. Our data also suggest that BqsS-BqsR may control biofilm decay by modulating biosynthesis of QS signals C4HSL and PQS (Pseudomonas quinolone signal).
Results
Deletion of bqsS and bqsR results in enhanced biofilm formation
Transposon mutagenesis was conducted to identify the genes of P. aeruginosa implicated in biofilm production. Screening of about 20,000 transposon mutants led to identification of fifteen mutants with altered biofilm phenotypes. While most of the biofilm mutants were owing to mutation of the previously identified genes (data not shown), a few mutants were found to contain transposon insertion in unreported genes. One mutant, in which PA2656 was disrupted by Marinar transposon insertion at the position 386 bp downstream the start codon ATG, was found to produce a significantly higher amount of biofilm than the wild-type strain mPAO1. PA2656 encodes a putative two-component sensor, containing a HAMP (Histidine kinases, Adenylyl cyclases, Methyl binding proteins, Phosphatases) domain, a phospho-acceptor domain, a histidine kinase-like ATPase domain, and a type I export signal peptide at N-terminal (http://v2.pseudomonas.com/) (). At the upstream of PA2656, the gene PA2657 was predicted to encode a response regulator that contains a CheY-type REC signal receiver domain at the N-terminal and an effector domain (reg_C) associated with DNA- and RNA-polymerase binding at the C-terminal (). Given their roles in biofilm development and QS as discussed below, these two genes were named as bqsS and bqsR, respectively. A BLAST search and gene context analysis showed that bqsS and bqsR, are highly conserved among Pseudomonas species, including P. mendocina, P. fluorescens, P. syringae, P. putida and P. entomophila, with an identity about 60%–80% at the amino acid level. In addition, other bacterial species, such as Nitrosococcus oceani, Alcanivorax borkumensis, Escherichia coli and Limnobacter sp. also contain homologues of BqsS and BqsR. None of these homologues has been previously characterized except the QseB of E. coli, which is a response regulator modulating bacterial cell motility and flagella biosynthesis.Citation19
To understand the role of BqsS-BqsR two-component system in biofilm development, we generated the bqsS and bqsR deletion mutants ΔbqsS and ΔbqsR, respectively, using P. aeruginosa mPAO1 as the parental strain. After grown in LB medium for 16 h at 37°C, biofilm mass was detected by crystal violet staining. Both ΔbqsS and ΔbqsR mutants showed enhanced biofilm formation compared with their parental strain ( and C). To further confirm the role of the BqsS-BqsR two-component system, the wild-type bqsS and bqsR genes were placed under the control of the lac promoter and introduced into the ΔbqsS and ΔbqsR mutants, respectively. The complemented strains were cultured under the same conditions as mPAO1 and its deletion mutants before determination of biofilm formation. The results showed the biofilm production by the complemented strain ΔbqsS(bqsS) and ΔbqsR(bqsR) was substantially decreased to a level even lower than that of the parental wild-type strain. Consistent with the notion that the response regulator BqsR acts at the downstream of the sensor BqsS, the data also showed that the strain ΔbqsS(bqsR), in which the response regulator gene bqsR was expressed in the sensor mutant ΔbqsS, produced much less biofilm ( and C). In contrast, expression of the sensor gene bqsS in the deletion mutant ΔbqsR was unable to rescue the biofilm phenotype of the response regulator mutant ( and C).
The BqsS-BqsR two-component system is involved in regulation of biofilm decay
To understand how the two-component system could influence biofilm formation, a time course analysis of biofilm formation was performed on these strains over a period of 32 h. The results showed that the major differences between the wild-type mPAO1 and the mutants were in biofilm accumulation and in extent of biofilm decay (). mPAO1 displayed a pattern of time-dependent accumulation and decline of biomass; in the first 16 h of incubation, the biomass of biofilms increased and reached a maximum amount at the time point of 16 h followed by a progressive decrease. In contrast, both ΔbqsS and ΔbqsR mutants, particularly the ΔbqsS mutant, produced abundant biofilms and displayed reduced biofilm decay over the course of the experiment. As expected, the biofilm development/decline patterns of the complemented stains were similar to the parental wild-type strain (). These data suggest that the increased biofilm mass in mutants ΔbqsS and ΔbqsR may be attributable to attenuated cell dispersion ability.
To investigate whether mutations in bqsS and bqsR might affect bacterial growth, which could influence the biomass of biofilm, we monitored bacterial growth using a Bioscreen growth apparatus. shows that the growth curves of the wild-type, deletion mutants and complemented strains were similar except that the mutant ΔbqsS displayed a slightly lower OD600 than other strains at the stationary phase, while the complemented strain ΔbqsS(bqsS) had a slightly higher OD600. Time course growth analysis of flask-cultured bacteria also showed similar results (data not shown). We further determined the total bacterial cell number in 24-h bacterial culture (biofilm-associated cells were mechanically dispersed by vortexing) using heterotropic plate counting assay, and found that the total bacterial cell numbers of five bacterial strains were similar in the range of 2.7–3.6 x 109 CFU/ml.
The wild-type supernatants reduce biofilm mass of the sensor mutant ΔbqsS
To investigate the molecular mechanism by which BqsS-BqsR regulates biofilm decay, a mix culture experiment was performed. The ΔbqsS mutant and wild-type mPAO1 were co-cultured in 2:1 and 1:1 ratio. After 24 h incubation in polystyrene tubes at 37°C, biofilm formation was quantified by crystal violet staining. The results showed that the amount of biofilms in the ΔbqsS-mPAO1 mix culture was significantly decreased in comparison with the ΔbqsS monoculture ().
The above data suggest that wild-type P. aeruginosa might produce an extracellular factor(s) that promotes biofilm decay. To test this possibility, ΔbqsS was inoculated in conditioned LB broth that contained 12.5% or 25% of filter-sterilized 24 h culture supernatants of the wild-type strain mPAO1. The bacteria were then grown at 37°C for 24 h before measurement of OD600 and biofilm formation. The results showed that inclusion of 12.5–25% of culture supernatants in conditioned LB medium had no obvious effect on growth rate of the mutant ΔbqsS (data not shown), but reduced mutant biofilm mass by up to 3-fold (). The results imply the presence of extracellular factor(s) in supernatants of P. aeruginosa that reduces biofilm biomass by either inhibiting biofilm formation or promoting biofilm decay.
The BqsS-BqsR two-component system controls rhamnolipid production
Several recent publications showed that rhamnolipids play a role in biofilm dispersion, detachment and biofilm structural development.Citation7,Citation8,Citation15 Therefore, rhamnolipid production, was investigated in mPAO1 and its derivatives by using thin-layer chromatography (TLC). The extracted rhamnolipids were separated on TLC plates. Purified rhamnolipids (JBR599; Jeneil Biosurfactant Co.,), which contain two predominant rhamnolipids, i.e., di-rhamnolipid and mono-rhamnolipid, were used as a control. TLC results showed that wild-type mPAO1 and the two complemented strains produced two rhamnolipids, whereas the mutants ΔbqsS and ΔbqsR yielded little or no rhamnolipids ().
We also found that deletion of bqsS and bqsR in wild-type strain mPAO1 decreased bacterial swarming motility by 27% and 19%, respectively, and the mutant phenotype was restored to the wild-type level by in trans expression of the corresponding wild-type gene. In contrast, deletion of the two-component system merely caused less than 5% change in swimming and twitching motility. These results appear to agree with the phenotype of decreased rhamnolipid production in the mutants, since rhamnolipids facilitate P. aeruginosa swarming motility by acting as biosurfactants.Citation20,Citation21 Given that the rhlAB operon is involved in production of rhamnolipids,Citation18 we investigated expression of the rhlAB operon by generation of a reporter gene prhlA'-lacZ, in which the lacZ gene was placed under the control of the rhlAB promoter. The β-galactosidase activities encoded by prhlA'-lacZ in strain mPAO1 and its derivatives were then determined. The assay results showed that expression of rhlAB in the mutants ΔbqsS and ΔbqsR were significantly reduced in comparison with their parental strain mPAO1 and the corresponding complemented strains (). The rhlAB expression pattern was similar to that of rhamnolipid production in these tested strains, suggesting that the BqsS-BqsR two-component system may influence expression of the genes encoding rhamnolipid biosynthesis and hence modulate production of rhamnolipids in P. aeruginosa.
The BqsS-BqsR two-component system influences production of C4HSL and PQS quorum sensing signals
Quorum sensing (QS) regulates expression of rhlAB and rhlC genes and production of rhamnolipids.Citation22–Citation24 It is intriguing to determine whether the BqsS-BqsR two-component system influences rhamnolipid production directly or via modulation of QS signaling. We therefore examined production of N-acyl-L-homoserine lactones (AHLs) QS signals in strain mPAO1 and its derivatives. AHLs were extracted from the overnight cultures grown at 37°C. The bioassay showed that strain mPAO1 and its mutants ΔbqsS and ΔbqsR produced similar levels of the long chain AHL signal 3OC12HSL (data not shown). However, decreased production of C4HSL was observed in the mutants ΔbqsS and ΔbqsR, especially the sensor mutant ΔbqsS, which produced over 7-fold less amount of C4HSL than its wild-type and complemented strain ().
Biosynthesis of the short chain C4HSL signal is not only regulated by the long chain signal 3OC12HSL but is also under positive control by the Pseudomonas quinolone signal (PQS), which is a QS signal linking the las and rhl QS systems.Citation25,Citation26 To determine whether BqsS-BqsR could influence PQS signaling, the signal extracts from strain mPAO1 and its derivatives were determined by TLC analysis following an established method.Citation27,Citation30 The results showed that the mutants ΔbqsS and ΔbqsR produced fewer PQS signals than strain mPAO1, whereas their complemented strains produced much more PQS than the wild-type (, left). Consistently, production of cytotoxin pyocyanin, which is positively regulated by PQS, was also reduced in ΔbqsS and ΔbqsR (). Similar to the pattern of PQS production, complemented strains produced higher levels of pyocyanin than wild-type, in particular the strain ΔbqsS(bqsS) (). The TLC analysis also detected anthranilate, which is a precursor of PQS,Citation28 in the wild-type and its deletion mutants; however, the precursor was hardly detectable in both complemented strains (). One plausible explanation is that overexpression of BqsS or BqsR in the complemented strains may facilitate conversion of anthranilate to PQS.
The reverse transcription polymerase chain reaction (RT-PCR) analysis confirmed the role of the BqsS-BqsR two-component system in modulation of transcriptional expression of pqsA and phnA, which are involved in PQS biosynthesis.Citation29 In both mutants ΔbqsS and ΔbqsR, the transcripts level of pqsA and phnA were decreased in comparison with mPAO1 and corresponding complemented strains ().
Exogenous addition of rhamnolipids, C4HSL or PQS reduce biofilm formation of the bqsS mutant to wild-type level
The above results suggest that the BqsS-BqsR two-component system may modulate QS-dependent rhamnolipid production and hence influence biofilm decay. We therefore tested whether exogenous addition of rhamnolipids and QS signal molecules to the ΔbqsS mutant could restore biofilm production to a wild-type level. Each test molecule was added to a final concentration of 50 and 100 µM, respectively, to LB liquid medium before inoculation of ΔbqsS. Quantification of biofilm 24 h after inoculation showed that addition of either rhamnolipids or C4HSL or PQS could reduce biofilm accumulation to a level similar to the wild-type mPAO1 (Suppl. Fig. S1A–C). To further assess the effect of these molecules on biofilm development at various growth stages, we monitored bacterial growth rate and biofilm mass at four time points, i.e., 6 h, 16 h, 24 h and 32 h after inoculation. At a final concentration of 50 mg/L (rhamnolipids) or 50 µM (C4HSL, PQS), the molecules neither affect the growth rate of ΔbqsS (Suppl. Fig. S1D), nor the initial biofilm formation (6 h) in comparison with the wild-type control, but significantly increased subsequent biofilm decay ().
Discussion
P. aeruginosa is able to grow either as planktonic- or biofilm-form and undergoes transition between these two lifestyles in response to environmental changes. However, the molecular mechanisms by which bacteria sense and respond to environmental changes and coordinate lifestyle transition are still poorly understood. In this study, we showed that null mutation of a novel multidomain sensor kinase BqsS and its cognate response regulator BqsR, in particular BqsS, resulted in enhanced biofilm formation and significantly reduced biofilm dispersion/detachment. Expression of the wild-type bqsS and bqsR in corresponding mutants restored the normal pattern of wild-type biofilm dispersion/detachment ( and C; ). In addition, overexpression of the response regulator BqsR in the sensor mutant ΔbqsS could restore its biofilm dispersion/detachment to the wild-type level ( and C). Cumulatively, these results have established an important role of the BqsS-BqsR two-component system in modulation of P. aeruginosa biofilm dispersion/detachment.
One of the key phenotypes regulated by the BqsS-BqsR two-component system is biosynthesis of rhamnolipids. Mutation of BqsS-BqsR resulted in significantly reduced production of rhamnolipids (), and expression of rhlAB (), whose products are involved in production of rhamnolipids.Citation18,Citation19 Rhamnolipids appear to have multiple roles in P. aeruginosa biofilm development, including promoting microcolony formation in the early growth phase and facilitating migration-dependent structural development in the late growth phase. It has been shown recently that P. aeruginosa rhlA mutants, deficient in synthesis of rhamnoslipids, are not capable of forming microcolonies in the initial phase of biofilm development.Citation19 In contrast, mutation of bqsS or bqsR appeared to significantly affect the biofilm development in the late growth phase but had little effect on the biofilm formation during early phase (). One plausible explanation is that BqsS-BqsR mutants were still able to produce a basal level of rhamnoslipids as evident from TLC analysis of rhamonoslipids and rhlAB expression analysis ( and B), which is sufficient to support initial biofilm formation.
Previous studies showed that rhamnolipids production is dependent on the short chain signal C4HSL-mediated QS system,Citation22,Citation23 and to a much lesser extent on the long chain signal 3OC12HSL-dependent QS system.Citation23 Our bioassay results showed that null mutation of either BqsS or BqsR did not affect 3OC12HSL accumulation in bacterial culture, but drastically reduced the production of C4HSL signals (). Chemical and genetic analysis also showed that the BqsS-BqsR two-component mutants produced substantially reduced PQS signals (). In light of our current results and the previous findings that C4HSL production is regulated positively by PQS signal,Citation30 we propose that the BqsS-BqsR two-component system may regulate P. aeruginosa biofilm decay through modulation of PQS, C4HSL and rhamnolipids production. The notion is strengthened by the findings that exogenous addition of either PQS, C4HSL or rhamnolipids at a physiological relevant concentration to the BqsS mutant was able to reduce the biofilm formation to a level similar to the wild-type strain mPAO1 (). Nevertheless, at this stage we have not yet tested the possibility that BqsS-BqsR could also regulate C4HSL biosynthesis in a PQS-independent manner. Neither would we rule out that other QS-dependent factors (apart from rhamnolipids) may also be implicated in the process of biofilm decay.
It is worthy noting that deletion of the sensor kinase gene bqsS resulted in much more significant phenotype changes, including biofilm decay ( and C; ), rhamnolipids production (), and C4HSL and PQS signal biosynthesis than the response regulator mutant ΔbqsR ( and C). One likely explanation is that the sensor BqsS may be able to communicate with more than one response regulators. Signal transduction cross-talk has been found between about 3% of non-cognate sensor/response regulator pairs out of 27 two-component systems in Escherichia coli.Citation31 The possibility that the sensor BqsS communicates with other response regulators in modulation of biofilm decay warrants further investigation.
It remains highly intriguing to determine the environmental signal(s) that activates the BqsS-BqsR two-component system. P. aeruginosa has more than 60 known or predicted two-component systems.Citation32 Among them, three sensor kinases, GacS,Citation33 RetSCitation34 and LadSCitation35 were found to affect biofilm formation by controlling the intracellular level of small regulatory RNAs. RetS and LadS are modular proteins sharing a similar domain architecture; both contain a N-terminal 7TMR-DISMED2 (7-transmembrane-receptor with diverse intracellular signaling modules extracellular domain 2), followed by a 7TMR-DISM_7TM domain (seven transmembrane segments found adjacent to 7TMR-DISM domain), and then C-terminal histidine kinase and response regulator receiver domains. The sensor RetS is larger than LadS because the former contains an additional response regulator domain at its C-terminal. Of particular note is that these two closely related sensors appear to play opposite roles on biofilm formation. The retS mutant showed a hyperadhesive phenotype and overproduction of biofilms,Citation34 whereas deletion of ladS resulted in loss of adherence and decreased biofilm formation.Citation34 Sequence alignment and domain analysis show that these two hybrid sensor proteins do not seem to share significant similarity with BqsS except the histidine kinase domain. The GacS-GacA two-component system has been reported to play a role in regulation of C4HSL production and biofilm formation in response to an unknown environmental signal.Citation33,Citation34,Citation36 The domain analysis and sequence alignment show that BqsS and GacS share the conserved HAMP-HisKA-HATPase domains with about 44% similarity but have little homology at the first 200 amino acids at the N-terminal, which may be implicated in sensing a signal ligand. Similar to the mutant ΔbqsR, the gacA mutant of P. aeruginosa strain PAO1 generates substantially less C4HSL signal,Citation33,Citation37 but the effect of the gacA mutation on biofilm formation in this strain has not been tested. In P. aeruginosa strain PA14, however, mutation of gacA does not seem to affect the C4HSL signal production but results in substantially decreased biofilm formation.Citation38 Despite this apparent inconsistency in modulating QS signal production, the role of GacS-GacA in positive regulation of biofim formation appears conserved in various bacterial species, including P. aeruginosa PA14,Citation38 P. aeruginosa PAK,Citation34 Pseudomonas sp. KL28,Citation39 and Erwinia chrysanthemi.Citation40 In all the cases, mutation of either GacS or GacA resulted in decreased biofilm formation, which is a sharp contrast to the negative regulatory role of the BqsS-BqsR system on biofilm development. Collectively, the available evidence seems to suggest that the BqsS-BqsR and GacS-GacA two-component systems may respond to different signals and use different mechanisms to influence biofilm formation.
In summary, we have demonstrated that BqsS-BqsR modulates biofilm decay by regulating biosynthesis of two QS signals, i.e., PQS and C4HSL. As these QS signal molecules are implicated in regulation of diverse biological functions,Citation18,Citation23,Citation26,Citation41 the BqsS-BqsR two-component system could also be an excellent model for further investigation on how bacteria may respond to environmental cues to adjust the QS-dependent social behaviors.
Materials and Methods
Bacterial strains, media, growth and motility assay
The P. aeruginosa strains and other bacteria used in this study are listed in . Unless otherwise indicated, bacteria were routinely grown at 37°C in Luria-Bertani broth (LB). Antibiotics were used when necessary at the following concentrations: carbenicillin, 300 µg/ml for P. aeruginosa, 200 µg/ml for Escherichia coli; tetracycline, 100 µg/ml for P. aeruginosa and 10 µg/ml for E. coli. For bacterial growth assay, overnight culture was diluted in LB broth and incubated in the Bioscreen C apparatus (Labsystems, Helsinki, Finland) at 37°C with moderate and continuous shaking. OD600 was measured every 30 min or otherwise indicated and each culture had 5 duplicates. Conditioned LB broth was obtained by addition of 12.5% to 25% of filter-sterilized overnight supernatants from strain mPAO1. Cell motility assays were performed as previously described.Citation42
DNA manipulation and deletion mutagenesis
The plasmids used in this study are listed in . To generate the bqsS and bqsR deletion mutants of P. aeruginosa, plasmids pEX-S and pEX-R were constructed as following: two PCR fragments flanking bqsS or bqsR were amplified from the genomic DNA of wild-type strain mPAO1. The 5′-and 3′-flanking regions of bqsS were amplified using the primer pair bqsS5f/bqsS5r (5′-AGCGAGCTCCAGGAGTGGCGTGGGCT-3′, 5′-GAAGAACTGTTAAGCCCTGGATCCGCTGGATCGACCTCATCC-3′), and bqsS3f/bqsS3r (5′-GATGAGGTCGATCCAGCGGATCCAGGGCTTAACAGTTCTTC-′3, 5′-GCTCTAGAGGAATCGGCCCAGGTCAG-3′), respectively. Similarly, the two flanking regions of bqsR were amplified using bqsR5f/bqsR5r (5′-AGCGAGCTCGAGCGTCCACGATACC-3′, 5′-GCTGGATCGACCTCATCCGGATCCTCAACCAGCAGCAAC-3′), and bqsR3f/bqsR3r (5′-GTTGCTGCTGGTTGAGGATCCGGATGAGGTCGATCCAGC-3′, 5′-GCTCTAGACCCATCCACTGCACACG-3′), respectively. The two flanking fragments of each gene were fused by overlap extension PCR. The resulted fusion PCR fragments contain the truncated bqsS and bqsR, in which a 1290-nt coding region (from 18–1308 bp) of bqsS and a 638-nt coding region (from 26–664 bp) of bqsR were deleted, respectively. After purification with QIAquick PCR purification kit (QIAGEN), the fusion fragments were digested with SacI and XbaI (the site is underlined in primer sequence) and separately cloned into the corresponding site of pEX18Ap vector.Citation43 The resultant constructs were introduced into E. coli S17-1 by electroporation and then mPAO1 by biparental mating. The generated bqsS and bqsR deletion mutants were confirmed by PCR.
For mutant complementation, the coding regions of the wild-type bqsS and bqsR genes were amplified from mPAO1 genomic DNA using primer pairs Sf/Sr (5′-GCTCTAGATGAGGTCGATCCAGCGGCG-3′, 5′-CACGGATCCTGTCGGCAAATGGTGAAGA-3′; XbaI and BamHI site underlined); Rf/Rr (5′-TGACAAGCTTATGCGGTTGCTGCTGGTTGA-3′, 5′-CACGGATCCGCTGGATCGACCTCATCC-3′; HindIII and BamHI site underlined), respectively. The PCR products were cloned at the downstream of lac promoter in the shuttle vector pUCP19 (ATCC 87110) digested by XbaI/BamHI (for bqsS) or HindIII/BamHI (for bqsR). The resultant constructs were mobilized into E. coli separately and sequenced before introducing to corresponding mutants as described in the previous section.
To construct the prhlA'-lacZ reporter plasmid, a 433 bp fragment corresponding to -377 to +56 bp relative to the translational start site of the rhlA gene was amplified from the genomic DNA of P. aeruginosa by PCR using the primer pair prhlAf 5′-TGACAAGCTTCATGCCTTTTCCGCCAAC-3′ and prhlAr 5′-CGGAATTCGACATGTACCCGCAGGCC-3′. The fragment was then digested with HindIII and EcoRI (underlined in the primer sequence) and then cloned in the same sites of pME2-lacZ, which was derived from pME6010.Citation44 The resultant constructs were used to transform P. aeruginosa by electroporation, and transformants were selected on LB agar plates containing relevant antibiotics.
RNA purification and RT-PCR analysis
Overnight culture of P. aeruginosa wild-type mPAO1 and its derivatives were diluted in LB broth and incubated at 37°C until the OD600 reached about 1.5. Total RNA samples were then isolated using the RNeasy miniprep kit (QIAGEN). Reverse transcription polymerase chain reaction (RT-PCR) was performed using QIAGEN OneStep RT-PCR Kit according to the manufacturer's protocol. The primer pairs used for pqsA and phnA are: pqsAF/pqsAR, (5′-TGATGCACAGCCTGCGCAAC-3′, 5′-CTTCCTCGATAGTGTGTCCT-3′); and phnAF/phnAR (5′-TTGCTGCTGGACATCGATCA-3′, 5′-AGCGATCGACCTTGAGCATG-3′), respectively.
β-galactosidase assay
For measurement of β-galactosidase activity, P. aeruginosa strains containing the prhlA'-lacZ construct were grown overnight in LB with shaking at 37°C. The starter cells were inoculated in LB at a ratio of 1:100, and then gown at the same temperature to an OD600 of 1.5. β-galactosidase activity was determined using a standard protocol.Citation45 Results are given as Miller units (MU) of β-galactosidase activity per OD600.
Biofilm formation assay and quantification
Biofilm formation assay was performed according to O'Toole and KolterCitation46 with minor modifications. Briefly, overnight bacterial cultures were diluted to the same concentration of OD600 = 0.002 with fresh LB broth. The diluted cultures (1 ml) were transferred to a 14-ml polystyrene tube (17 x 100 mm; FALCON, 352057) and incubated at 37°C with shaking at 210 rpm for a period as stated. Bacterial cultures were carefully removed for measurement of OD600. The bacterial cells bound to the wall of the tubes (biofilms) were stained with 0.1% crystal violet (Sigma) for 15 min at room temperature and the tubes were then rinsed several times with water. The tubes were air-dried at room temperature and then photographed. For quantification, the attached cells (biofilms) were suspended in 2 ml of 75% ethanol. The absorbance at 595 nm was measured with a spectrophotometer. Each experiment was repeated at least three times.
Preparation of rhamnolipids and TLC assay
Bacterial cells were removed from the cultures grown at 37°C for 24 h by centrifugation, and the supernatant was adjusted to pH 2 with concentrated HCl. Rhamnolipids were then extracted from the acidified supernatants (1 ml) twice with an equal volume of chloroform-ethanol (2:1, v:v) mixture. The pooled organic phases were evaporated to dryness, and the remaining residues were dissolved in 0.5 ml of methanol. After air dry, each sample was resuspended in 20 µl of menthol. An aliquot of each sample (5 µl) was spotted and analyzed by thin-layer chromatography (TLC; silica gel 60 plates, F254; Merck, Darmstadt, Germany) with a mobile phase consisting of chloroform-methanol-water (65:15:2 by volume). Commercial rhamnolipids (JBR599; Jeneil Biosurfactant Co., LCC) were used as a standard control in TLC analysis.
Pyocyanin, AHL and PQS extraction and assay
P. aeruginosa strains were grown in LB or Pseudomonas medium ACitation47 (for pyocyanin) broth for 24 h at 37°C with shaking. Pyocyanin was extracted from 5 ml of culture supernatant with a 3-ml volume of chloroform and the chloroform phase was extracted with 0.2 N HCl. The OD520 of the aqueous phase was measured. The AHL signals were extracted from 10 ml of supernatants with an equal volume of acidified ethyl acetateCitation26 by vigorous vortexing, followed by centrifugation. The organic phase was transferred to a fresh tube and dried to completion. The extracted compounds were dissolved in 100 µl methanol for bioassay. Long chain AHL signal 3OC12HSL were assayed by plate diffusion assay as described.Citation48 For detection of C4HSL, Chromobacterium violaceum CV026Citation49 was used as an indicator strain and LB agar medium was used for bioassay. Bioassay plates were incubated at 28°C for 40 h. The relative amounts of C4HSL were quantified as described.Citation48,Citation50 PQS extraction and assay were conducted as described by Diggle et al.Citation30 For each sample, 5-µl extracts were spotted onto a Silica 60 F254 plate (10 x 20 cm; Merck), along with synthetic PQS standard. TLC plates were soaked for 30 min in 5% KH2PO4 and activated at 100°C for 1 h before use. Chromatography was performed with a solvent mixture of dichloromethane-methanol (95:5). Upon completion, the plates were air-dried and visualized under UV light and photographed using AlphaImager (Alpha Innotech). For determination of anthranilate, TLC plates were left for about 24 h at room temperature until yellow spots became visible.Citation27
Figures and Tables
Figure 1 Null mutation of the BqsS-BqsR two-component system results in enhanced biofilm formation in P. aeruginosa strain mPAO1. (A) Genetic organization and domain structures of the sensor kinase BqsS and the response regulator BqsR. Gene orientation is indicated by arrow. Domain structure prediction was done using the SMART program (http://smart.emblheidelberg.de/). Symbol: REC, cheY-homologous receiver domain: reg_C: response regulator receiver domain; SP, signal peptide; HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins, phosphatases domain; HisKA, phospho-acceptor domain; HATPase, histidine kinase-like ATPase domain. (B) Visualization of biofilm formation on the walls of the polystyrene tubes by crystal violet staining. (C) Quantification of biofilm formation. The data shown are the means of triplicates and the standard deviation (SD) is shown by error bar. The following bacterial strains were used in this experiment: mPAO1 (mP), the bqsS deletion mutant ΔbqsS (ΔS), the bqsR mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], ΔbqsS(bqsR) [ΔS(R)], ΔbqsR(bqsR) [ΔR(R)] and ΔbqsR(bqsS) [ΔR(S)].
![Figure 1 Null mutation of the BqsS-BqsR two-component system results in enhanced biofilm formation in P. aeruginosa strain mPAO1. (A) Genetic organization and domain structures of the sensor kinase BqsS and the response regulator BqsR. Gene orientation is indicated by arrow. Domain structure prediction was done using the SMART program (http://smart.emblheidelberg.de/). Symbol: REC, cheY-homologous receiver domain: reg_C: response regulator receiver domain; SP, signal peptide; HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins, phosphatases domain; HisKA, phospho-acceptor domain; HATPase, histidine kinase-like ATPase domain. (B) Visualization of biofilm formation on the walls of the polystyrene tubes by crystal violet staining. (C) Quantification of biofilm formation. The data shown are the means of triplicates and the standard deviation (SD) is shown by error bar. The following bacterial strains were used in this experiment: mPAO1 (mP), the bqsS deletion mutant ΔbqsS (ΔS), the bqsR mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], ΔbqsS(bqsR) [ΔS(R)], ΔbqsR(bqsR) [ΔR(R)] and ΔbqsR(bqsS) [ΔR(S)].](/cms/asset/076cc389-a93a-4a32-bb72-5f50c1e98081/kcib_a_10906717_f0001.gif)
Figure 2 Time course assay of biofilm development. (A) Quantification of biofilm formation at different time points. P. aeruginosa strains were grown in LB medium in 14-ml polystyrene tubes and the amounts of biofilm mass at different time points as indicated were quantified. Assays were performed in triplicates and the data were the mean values ± SD. (B) Bioscreen analysis of the bacterial growth. The data were the mean values of 5 replicates. The following bacterial strains were used in this experiment: mPAO1 (mP), mutant ΔbqsS (ΔS), mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], and ΔbqsR(bqsR) [ΔR(R)].
![Figure 2 Time course assay of biofilm development. (A) Quantification of biofilm formation at different time points. P. aeruginosa strains were grown in LB medium in 14-ml polystyrene tubes and the amounts of biofilm mass at different time points as indicated were quantified. Assays were performed in triplicates and the data were the mean values ± SD. (B) Bioscreen analysis of the bacterial growth. The data were the mean values of 5 replicates. The following bacterial strains were used in this experiment: mPAO1 (mP), mutant ΔbqsS (ΔS), mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], and ΔbqsR(bqsR) [ΔR(R)].](/cms/asset/4fd77939-4dc9-4390-b1b9-d712ba66aa9d/kcib_a_10906717_f0002.gif)
Figure 3 Effect of co-culture and wild-type supernatants on biofilm formation by mutant ΔbqsS. (A) Biofilm formation by co-cultured bacteria. The mutant ΔbqsS (ΔS) was mixed with wild-type mPAO1 (mP) in 2:1 or 1:1 ratio and biofilms were quantified after incubation for 24 h. (B) Biofilms formed in conditioned medium. Wild-type mPAO1 and mutant ΔbqsS were cultured in conditioned medium which contained 12.5% or 25% of the supernatants of mPAO1 in LB broth. The data are the means of triplicates and SD is shown by error bar.
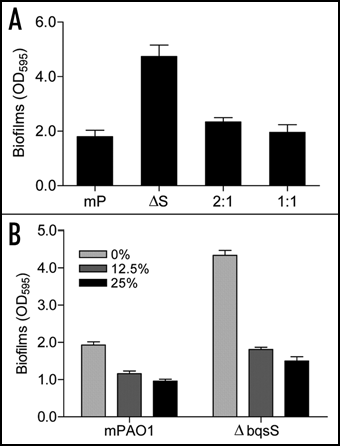
Figure 4 Effect of bqsS-bqsR mutation on rhamnolipid production and rhlA expression. (A) TLC plate assay of rhamnolipids production. P. aeruginosa strains described in were grown at 37°C for 24 h, and rhamnolipids were extracted from the supernatants for TLC analysis using standard rhamnolipids (Rh) as a positive control and the extracts from lasI and bqsS double mutant (IS) was used as negative control. Two predominant rhamnolipids, mono-rhamnolipids (mono-RL) and di-rhamnolipids (di-RL) were indicated by arrows. (B) The rhlA'-lacZ fusion gene expression assay. Different bacterial strains containing the prhlA-lacZ construct were grown in LB broth at 37°C to OD600 of 1.5 and the cells were then collected and assayed for β-galactosidase activity. The data were the means of triplicate with standard deviations. The bacterial strains used were indicated in .
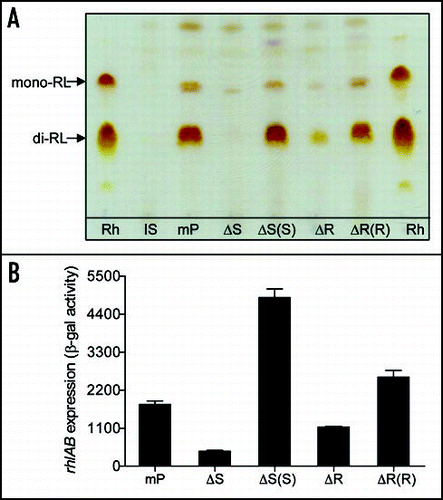
Figure 5 Effect of bqsS and bqsR deletion on QS signal production. (A) C4HSL production. C4HSL was extracted from the supernatants of P. aeruginosa strains and assayed by using indicator strain Chromobacterium violaceum CV026. Synthetic C4HSL was used as a positive control. Quantification was performed as describedCitation48,Citation50 and the data were presented as relative percentages to wild-type. (B) Pyocyanin production by P. aeruginosa strains as indicated. (C) TLC analysis of PQS (left) and anthranilate (right). PQS and anthranilate were extracted from supernatants of P. aeruginosa strains. The extracts were separated on TLC plates and the plates were visualized under UV (left) or natural light (right). Synthetic PQS (37 nmole) and anthranilate (25 nmole) were spotted separately as controls. (D) RT-PCR analysis of the transcript levels of pqsA and phnA. The proC gene, which is constitutively expressed, was amplified under the same conditions as an internal loading control.
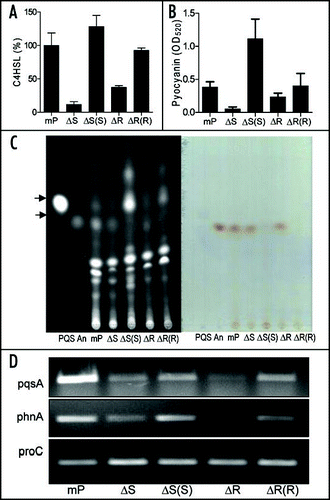
Figure 6 Effect of exogenous addition of rhmnolipids, C4HSL and PQS on biofilm formation by mutant ΔbqsS. P. aeruginosa strain mPAO1 (mP) was grown in LB medium as a control, and ΔbqsS (ΔS) was inoculated in the same medium with or without rhamnolipids (Rh, 50 µg/ml) or C4HSL (50 µM) or PQS (50 µM). The bacteria were grown for 6, 16, 24 and 32 h before quantification of biofilm formation. The data were the means of triplicates with standard deviation.
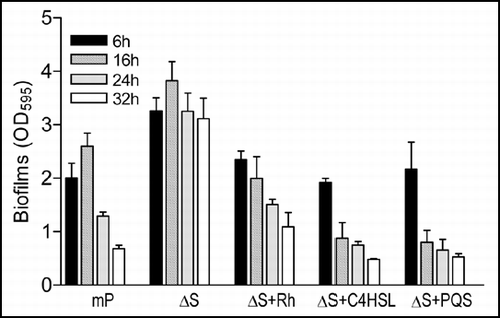
Table 1 Bacterial strains and plasmids used in this studyTable Footnotea
References
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol 1995; 49:711 - 745
- Quinn JP. Pseudomonas aeruginosa infections in the intensive care unit. Semin Respir Crit Care Med 2003; 24:61 - 68
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999; 284:1318 - 1322
- O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol 2000; 54:49 - 79
- Stoodley P, Cargo R, Rupp CJ, Wilson S, Klapper I. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J Ind Microbiol Biotechnol 2002; 29:361 - 367
- Klausen M, Gjermansen M, Kreft JU, Tolker-Nielsen T. Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol Lett 2006; 261:1 - 11
- Schooling SR, Charaf UK, Allison DG, Gilbert P. A role for rhamnolipid in biofilm dispersion. Biofilms 2004; 1:91 - 99
- Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 2005; 57:1210 - 1223
- Bollinger N, Hassett DJ, Iglewski BH, Costerton JW, McDermott TR. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J Bacteriol 2001; 183:1990 - 1996
- Hunt SM, Werner EM, Huang B, Hamilton MA, Stewart PS. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl Environ Microbiol 2004; 70:7418 - 7425
- Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 2004; 186:7312 - 7326
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature 2002; 417:552 - 555
- Allison DG, Ruiz B, SanJose C, Jaspe A, Gilbert P. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol Lett 1998; 167:179 - 184
- Boyd A, Chakrabarty AM. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl Environ Microbiol 1994; 60:2355 - 2359
- Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol 2007; 189:2531 - 2539
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998; 280:295 - 298
- Heydorn A, Ersbøll B, Kato J, Hentzer M, Parsek MR, Tolker-Nielsen T, Givskov M, Molin S. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl Environ Microbiol 2002; 68:2008 - 2017
- Ochsner UA, Fiechter A, Reiser J. Isolation, characterization and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem 1994; 269:19787 - 19795
- Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 2002; 43:809 - 821
- Deziel E, Lepine F, Milot S, Villemur R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 2003; 149:2005 - 2013
- Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 2000; 182:5990 - 5996
- Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 1995; 92:6424 - 6428
- Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorumsensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 1997; 179:5756 - 5767
- Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberon-Chavez G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol 2001; 40:708 - 718
- McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol 2000; 182:2702 - 2708
- Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 1999; 96:11229 - 11234
- Baysse C, Cullinane M, Denervaud V, Burrowes E, Dow JM, Morrissey JP, Tam L, Trevors JT, O'Gara F. Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology 2005; 151:2529 - 2542
- Bredenbruch F, Nimtz M, Wray V, Morr M, Muller R, Haussler S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J Bacteriol 2005; 187:3630 - 3635
- Diggle SP, Cornelis P, Williams P, Camara M. 4-quinolone signalling in Pseudomonas aeruginosa: old molecules, new perspectives. Int J Med Microbiol 2006; 296:83 - 91
- Diggle SP, Winzer K, Chhabra SR, Worrall KE, Camara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol 2003; 50:29 - 43
- Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem 2005; 280:1448 - 1456
- Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many?. Trends Microbiol 2000; 8:498 - 504
- Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide and lipase. Mol Microbiol 1997; 24:309 - 319
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 2004; 7:745 - 754
- Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA 2006; 103:171 - 176
- Dubuis C, Rolli J, Lutz M, Defago G, Haas D. Thiamine-auxotrophic mutants of Pseudomonas fluorescens CHA0 are defective in cell-cell signaling and biocontrol factor expression. Appl Environ Microbiol 2006; 72:2606 - 2613
- Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J Bacteriol 2006; 188:6026 - 6033
- Parkins MD, Ceri H, Storey DG. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol 2001; 40:1215 - 1226
- Choi KS, Veeraragouda Y, Cho KM, Lee SO, Jo GR, Cho K, Lee K. Effect of gacS and gacA mutations on colony architecture, surface motility, biofilm formation and chemical toxicity in Pseudomonas sp. KL28. J Microbiol 2007; 45:492 - 498
- Yang S, Peng Q, Zhang Q, Yi X, Choi CJ, Reedy RM, Charkowski AO, Yang CH. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol Plant Microbe Interact 2008; 21:133 - 142
- Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 2002; 184:6472 - 6480
- Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 2000; 97:4885 - 4890
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998; 212:77 - 86
- Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'Gara F, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact 2000; 13:232 - 237
- Sambrook JF, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual 1989; Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 1998; 30:295 - 304
- King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 1954; 44:301 - 307
- Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA 2000; 97:3526 - 3531
- McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GS, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997; 143:3703 - 3711
- Zhang HB, Wang LH, Zhang LH. Coico R, Stevenson JQ B. Detection and analysis of quorum-quenching enzymes against acyl homoserine lactone qurom-sensing signals. Current Protocols in Microbiol 2007; John Wiley and Sons, Inc.
- Simon R, Priefer U, Puhler A. A broad range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technol 1983; 1:784 - 791