Abstract
Eisosomes are punctate structures located in the cytoplasmic side of the cell membrane of ascomycetes. In Saccharomyces cerevisiæ they coincide topologically with and are necessary for the organisation of specific membrane domains. The eisosomal proteins are universally and quite strictly conserved in the sub-phylum, however this evolutionary conservation is in apparent contradiction with an elusive functional significance. The comparative analysis of the eisosomes of S. cerevisiae and Aspergillus nidulans reveal striking differences in the assembly and developmental fate of these structures between these two model organisms.
In 2006, a new organelle of Saccharomyces cerevisiae was described, and named “eisosome.”Citation1 Eisosomes underlie a compartment (MCC), which corresponds to furrows of the plasma membrane and comprises membrane proteins including three characterised transporters. Two of these (Can1 and Tat2) are amino acid transporters, but no other proteins of the highly conserved YAT family seem to share this localisation. Similarly, of the three proteins of the Fur family, only one, Fur4, is localised to MCC.Citation2 Nothing is known as to why among closely related proteins some localise to MCC and others do not. In A. nidulans, all characterised transporters, belonging to widely different families, show a continuous rather than a punctate pattern,Citation3 (including a transporter of unknown function of the GPR1/FUN34/yaaH familyCitation4). The orthologue of Fur4 and other members of this family have been characterised in A. nidulansCitation5,Citation6 however, their membrane localisation has not been determined.
In S. cerevisiae two homologous non-membrane proteins, Pil1 and Lsp1, compose eisosomes. We don't know how these proteins are anchored to the membrane. In the three sub-phyla of the ascomycetes (Saccharomycotina, Taphrinomycotina and Pezizomycotina) two homologues of Pil1 and Lsp1 are extant in the genomes, arising from three cognate independent duplications. In the Pezizomycotina one tight clade (PilA) is highly similar to the two S. cerevisiae proteins, while a second one (PilB), shows a much higher degree of intra-clade divergence.Citation3 This is illustrated in for the homologues present in the Aspergilli. Pil1 (and presumably Lsp1) is a phosphorylated proteinCitation7,Citation8 and PilA of A. nidulans show a higher conservation of putative phosphorylation sites than PilB (). shows that while proteins of the PilA clade (and the homologue proteins in the Saccharomycetales) comprise a predicted one coil-coiled domain, a second one is always predicted for the proteins of the PilB clade. We have shown that the eisosomes of the conidiospores of A. nidulans are composed by both PilA and PilB, and thus are similar to the eisosomes of S. cerevisiae, while the mycelial structures only contain PilA. The conidiation of A. nidulans has been compared to S. cerevisiae budding, but while in this yeast eisosomes assemble in a polarised wave from the neck to the tip of the bud,Citation9 the conidial eisosomes of A. nidulans assemble in already mature, non dividing spores (Fig. 5 of ref. Citation3). Particularly baffling is the presence of cytoplasmic, undegraded, presumably free PilB in germlings and mycelia. It is tempting to correlate the differences in phosphorylation sites and the presence of an additional coil-coiled domain with the different developmental fates of PilA and PilB.
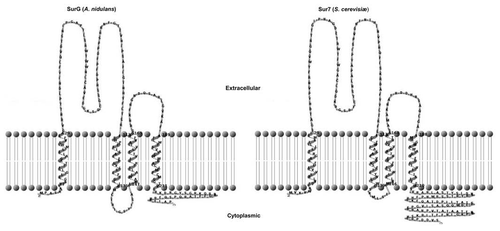
Historically, the first membrane protein associated with eisosomes to be investigated was Sur7.Citation1,Citation10 The characterised S. cerevisiae, Candida albicans and A. nidulans (SurG) homologues have four predicted transmembrane domains. This has been confirmed experimentally for Sur7.Citation11 The predicted topology of the S. cerevisiae and A. nidulans proteins is shown in . Strict homologues with four predicted transmembrane domains can be found in all ascomycete genomes, in variable number, while more divergent proteins with 3–5 predicted membrane domains can be found in the different sequenced genomes (see Sup. Fig. 1 of ref. Citation3). However, neither Sur7 nor its A. nidulans orthologue SurG are necessary for the punctate appearance of Pil1/PilA. Pil1 has a crucial role in S. cerevisiae, in its absence, no punctate structures of Lsp1 or Sur7 can be seen, however, this role is not identical in the conidiospores of A. nidulans, where in the absence of PilA distinct patches of PilB (but not of SurG) can still be seen in the periphery of the cell.
A second membrane protein, Nce102 seems to have a more crucial role in eisosome assembly.Citation2,Citation12 A model of membrane furrow formation proposes that Pil1 determines the sites where Sur7 and then Nce102 bind, and subsequently Nce102 recruits other membrane proteins.Citation13 However, as the absence of Nce102 results in a strikingly altered distribution of Pil1, this sequential sequence of events cannot be the whole story. Recent data strongly suggest that Nce102 is functionally conserved among the ascomycetes.Citation11 Strict homologues are present in all the Aspergilli. The 6 carboxy-terminal residues are essential for Nce102 function,Citation11 and indeed these are PTISVQ in S. cerevisiae PVMSVQ in Schizosaccharomyces pombe and PSMSVQ in A. nidulans. Further work should determine whether the A. nidulans homologue is active in the assembly of eisosomes on conidia and/or mycelia of A. nidulans.
The universal conservation among the ascomycetes of both the proteins of eisosomes and those associated with the cognate membrane furrows is indeed a paradox. No strong growth or developmental phenotypes are observed in deletions of the cognate genes in either S. cerevisiae or for those investigated by us in A. nidulans. Eisosomes were first described as sites of endocytosis, however, further work show that Can1 is actually released from MCCs when endocytosis is elicited and that endocytosis markers do not co-localise with eisosomes.Citation2 The work in A. nidulans does not support (but given the limited number of cargoes checked, cannot exclude) any role in endocytosis. Thus we have a complex and strongly conserved cellular apparatus, which has even been called an organelle, without an obvious physiological function. It will be most interesting to know what is the developmental distribution and eventual role of eisosomal proteins in other filamentous fungi, which have, like Aspergillus nidulans complex asexual and sexual cycles.
Abbreviations
| MCC | = | membrane compartment occupied by Can1 |
Figures and Tables
Figure 1 Phylogeny and Conservation of Pil-like proteins. (A) An unrooted maximum likelihood tree (for detail of methods see legend of Fig. 1 of ref. Citation3) showing the PilA and PilB clades of the Aspergilli. For reasons of clarity the systematic names of the autocalled genes are not shown in the tree, and are reported here. For the PilA clade: Afu: A. fumigatus, Afu6g07520; Nfi: Neosartoria fischeri, NFI_053190; Aor: A. oryzæ, AO090005001548; Afl: A. flavus, AFL2G_01442.2; Ate: A. terreus ATG _03673.1; Acl: A. clavatus ACL_081680; Ang: A. niger, An07g08890. For the PilB clade the same abbreviations are used and cognate systematic names are: Afu6g08320, NFI_053960, AO090003000094, AFL2G_02857.2, ATEG _05825.1, ACLA_082350, An11g01750. PilA and PilB denote the paralogues of A. nidulans (see text). The PilA and Lsp1 proteins of S. cerevisiae are also included in the tree. For reasons of space the aLRT branch support values are not shown within the PilA cluster, the lowest value being 0.74. (B) Eisosomal proteins in the Aspergilli. Comparison of the Pil1p, Lsp1 proteins of S. cerevisae and the PilA and PilB proteins of A. nidulans. Alignment carried out with MUSCLE, shading with Boxshade. Triangles, phosphorylation sites reported by Luo et al.Citation7 inverted triangles phosphorylation sites reported by Walther et al.Citation8
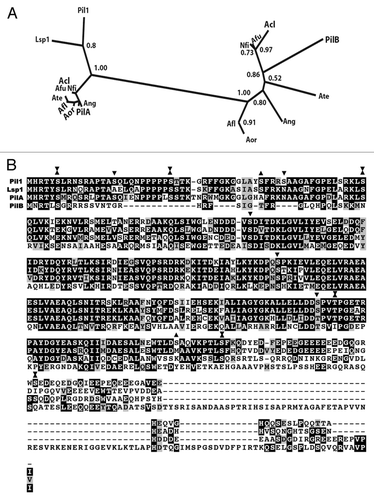
Figure 2 Proteins of the PilB clade have two putative coiled-coil domains. PilA and PilB proteins of the species of the Pezizomycotina included in the phylogeny of Figure 1 of reference Citation3 were independently aligned with T-Coffee. The alignments were scanned for coiled-coil domains with http://toolkit.tuebingen.mpg.de/pcoils. Size of window used by the programme: Green: 14 residues, blue: 21 residues, red: 28 residues. For the PilA clade, as a result of their high sequence identity, predictions for individual proteins do not differ from the prediction for the six aligned proteins. For the PilB clade, the sequence divergence leads to somewhat different individual predictions. The prediction for the coiled-coil domains of A. nidulans PilB is shown in the bottom panel.
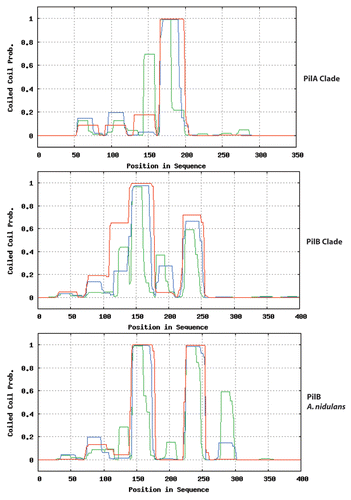
Figure 3 Predicted topology of SurG (A. nidulans) and Sur7 (S. cerevisiae). Graphic representation of the secondary structures of these proteins obtained with TMRP res2D (bioinformatics.biol.uoa.gr/TMRPres2D/index.jsp) based on the result from the TMHMM topology prediction tool (www.cbs.dtu.dk/services/TMHMM/).
Addendum to:
References
- Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. Eisosomes mark static sites of endocytosis. Nature 2006; 439:998 - 1003
- Grossmann G, Malinsky J, Stahlschmidt W, Loibl M, Weig-Meckl I, Frommer WB, et al. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol 2008; 183:1075 - 1088
- Vangelatos I, Roumelioti K, Gournas C, Suárez T, Claudio Scazzocchio C, Sophianopoulou V. Eisosome organisation in the filamentous ascomycete Aspergillus nidulans. Eukaryotic Cell 2010; 9:1441 - 1454
- Flipphi M, Robellet X, Dequier E, Leschelle X, Felenbok B, Vélot C. Functional analysis of alcS, a gene of the alc cluster in Aspergillus nidulans. Fungal Genet Biol 2006; 43:247 - 260
- Amillis S, Hamari Z, Roumelioti K, Scazzocchio C, Diallinas G. Regulation of expression and kinetic modelling of substrate interactions of a uracil transporter of Aspergillus nidulans. Mol Membr Biol 2007; 24:206 - 214
- Hamari Z, Amillis S, Drevet C, Apostolaki A, Vágvölgyi C, Diallinas G, et al. Convergent evolution and orphan genes in the Fur4p-like family and characterization of a general nucleoside transporter in Aspergillus nidulans. Mol Microbiol 2009; 73:343 - 357
- Luo G, Gruhler A, Liu Y, Jensen ON, Dickson RC. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J Biol Chem 2008; 283:10433 - 10444
- Walther TC, Aguilar PS, Fröhlich F, Chu F, Moreira KE, Burlingame AL, et al. Pkh-kinases control eisosome assembly and organization. EMBO J 2007; 26:4946 - 4955
- Moreira KE, Walther TC, Aguilar PS, Walter P. Pil1 controls eisosome biogenesis. Mol Biol Cell 2009; 20:809 - 818
- Alvarez FJ, Douglas LM, Rosebrock A, Konopka JB. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol Biol Cell 2008; 19:5214 - 5225
- Loibl M, Grossman G, Stradalova V, Klingl A, Rachel R, Tanner W, et al. C terminus of Nce102 determines the structure and function of microdimains in the Saccharomyces cerevisiae plasma memebrane. Eukaryotic Cell 2010; 9:1184 - 1192
- Fröhlich F, Moreira K, Aguilar PS, Hubner NC, Mann M, Walter P, et al. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J Cell Biol 2009; 185:1227 - 1242
- Malinski J, Opekarova M, Tanner W. The lateral compartmentation of the yeast plasma membrane. Yeast 2010; 27:473 - 478