Abstract
We recently described a novel bet-hedging mechanism in which the bacterium Sinorhizobium meliloti responds to starvation by forming two discrete cell types via cell division. The old-pole daughter cell retains most of the resource, polyhydroxybutyrate (PHB) and is capable of surviving long-term starvation, while the low-PHB, new-pole daughter cell is capable of quickly resuming growth when starvation ends. Here we present additional data showing that the high-PHB, old-pole cells are similar to bacterial persisters, characterized by metabolic dormancy and antibiotic tolerance. Using two independent methods, we generated clonal populations of S. meliloti that varied in the frequency of the high- and low-PHB phenotypes, and then challenged these populations with ampicillin. Populations containing more high-PHB cells were significantly more antibiotic-tolerant. In a separate experiment, we used GFP fluorescence as a marker of overall metabolic activity. After 24 hours of starvation, new-pole cells were 64% brighter than their old-pole sister cells, demonstrating that the divergence in metabolic rate is rapid.
We recently demonstrated that, when starved, the bacterium Sinorhizobium meliloti uses energy and carbon stored in the polyester poly-3-hydroxybutyrate (PHB) to divide, producing one high-PHB cell and one or two low-PHB cells.Citation1,Citation2 High-PHB cells, but not low-PHB cells, are capable of surviving more than a year of starvation, but take longer to resume growth once starvation ends.Citation1 The latter result suggests that the high-PHB phenotype may be dormant. In addition to conserving stored resources, dormant bacteria can gain resistance to environmental stresses, like antibiotics and heavy metals, that can kill metabolically active cells.Citation3,Citation4 Most clonal populations of bacteria contain a small proportion of these dormant, antibiotic-tolerant ‘persister’ cells.Citation5 After starvation and differentiation into high and low-PHB phenotypes, are high-PHB cells entering a state of persistence?
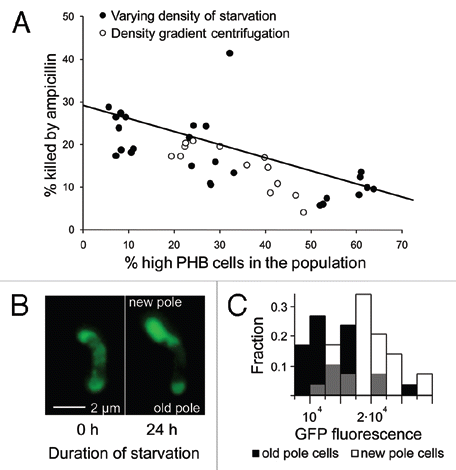
We examined this by generating groups of genetically-uniform, starved S. meliloti with varying fractions of high-PHB-phenotype cells, and then exposed these populations to ampicillin at 500 µgmL−1 for 90 minutes. Ampicillin is a broad spectrum β-lactam antibiotic that only affects actively growing bacteria; as a result it is a widely-used screen for persisters.Citation3,Citation5–Citation7 We used two independent methods to generate variation in the frequency of high-PHB cells. First, density gradient centrifugation was used to fractionate five populations of starved cells (with bimodal PHB content) into low (≤1.109 gmL−1), medium (1.119-1.109 gmL−1) and high (1.138-1.119 gmL−1) density as previously described in reference Citation2; fractions with a higher buoyant density contained more high-PHB cells. Second, we starved S. meliloti at high (5 × 106), medium (5 × 105) and low (105) density for three days; cells starved at lower densities reproduced more and formed populations with fewer high-PHB cells. Viability was assayed flow cytometrically for both experiments as previously described1 except that we used viability stains propidium iodide in the centrifugation experiment and YO-PRO-1 in the density-of-starvation experiment. Both viability stains are functionally equivalent ways of detecting the failure of dead cells to exclude DNA-binding stains.Citation1 Populations with mostly high-PHB rhizobia survived antibiotic exposure significantly better than those with mostly low-PHB rhizobia (; p < 0.0001, n = 41, r2 = 0.46, linear regression). S. meliloti strain 1021 is normally killed by ampicillin, but the high-PHB phenotype is apparently tolerant, consistent with results for other bacterial persisters.
We next tested for persistence using an independent method. Because persisters are metabolically dormant, protein production is reduced relative to growing cells. Therefore, bacteria that are engineered to constitutively produce GFP glow less brightly when they become persisters.Citation5 We therefore examined GFP fluorescence in S. meliloti strain pDG71, engineered to constitutively produce GFP.Citation8,Citation9 We followed the fate of initially high-PHB cells that were affixed to a coverslip and incubated in starvation media using methods reported previously in ref. Citation1. Mean GFP fluorescence per unit area was assessed for 30 dividing cells after 24 hours of starvation. Compared to their old-pole parents, new-pole cells were 65% brighter ( and C; t = 8.35, n = 30, p < 0.0001, matched-pair t-test).
To summarize, high-PHB cells respond more slowly to exogenous resources,Citation1 are more antibiotic-tolerant, and are less GFP fluorescent than low-PHB cells, demonstrating that they are in a state of persistence. The low metabolic rate (persister phenotype) of the old-pole cell would complement its retention of a greater share of PHB during cell division, further enhancing the greater longevity of old-pole cells that is key to their bet-hedging strategy. The putative role of the high-PHB phenotype is longevity; old-pole, high-PHB cells can survive more than a year of starvation to resume growth or nodulation if conditions are slow to change. Persistence is an effective way to increase longevity if mortality from antibiotics and other stresses that kill metabolically-active cells is high. The cost of persistence, a missed opportunity to nodulate or consume exogenous resources if conditions improve suddenly, is ameliorated through individual-level bet hedging. Because individual S. meliloti produce both persistent and non-persistent cells when starved, resources that cannot be used by a dormant cell may instead be used by its nearby metabolically-active clonemate.
Bacterial persistence is widely thought of as an insurance policy taken out by a genotype (expressed by a tiny fraction of the clonal population) as a hedge against catastrophe.Citation10 Recent work, however, suggests a more dynamic role for persistence in microbial life-history evolution. Dorr et al.Citation11 have shown that persistence can be induced by exposure to the antibiotic ciproflaxin. This type of phenotypic plasticity allows for much higher survival than ‘catastrophe insurance’, where only a tiny fraction of the population survives. Theoretical work has proposed alternative explanations for the evolution of persistence, including reduced competition with kin for limited resourcesCitation12 or as a side-effect of senescence,Citation13 but these hypotheses await experimental confirmation. Our findings, that persistence can be utilized in an individual-level bet-hedging strategy that is triggered in response to stress, adds to the diversity of life-history strategies that employ persistence.
Abbreviations
| PHB | = | poly-3-hydroxybutyrate |
| GFP | = | green fluorescent protein |
| PI | = | propidium iodide |
Figures and Tables
Figure 1 High-PHB old-pole cells enter a persistent state. (A) Populations of Sinorhizobium meliloti that varied in the percentage of high vs. low-PH B cells were generated via density gradient centrifugation or by starving rhizobia at varying densities. These rhizobia were then challenged with ampicillin and assayed for mortality (PI or YO-PRO-1 uptake) by flow cytometry. Populations containing a greater fraction of high-PHB cells were less susceptible to ampicillin. There was no significant difference in linear regression slopes (t = 0.8, p = 0.43, n = 41, ANCOVA) or intercepts (t = 1, p = 0.32, n = 41, ANCOVA) for the two methods of manipulating the frequency of high-PH B cells, so a single linear regression was fit to the data. High-PH B old pole cells are less GFP fluorescent (B and C), indicating lower metabolic activity. (B) An initially high-PH B cell dividing during starvation produces a bright, GFP fluorescent new-pole cell and a dim old-pole cell. To increase brightness and lessen background fluorescence, this image was transformed with the ‘Colors; Levels’ command in GIMP v. 2.6. Both left and right panels were transformed identically. This transformation made the cells easier to see and did not qualitatively affect the image. (C) After 24 hours of starvation, GFP fluorescence intensity is 50.8% higher in new-pole cells (t = 5.29, n = 60, p < 0.0001, two-sided t-test). Gray bars represent overlap in new and old-pole distributions. Brightness units are average color depth (ranging from 0–65536) of the green channel from a 16-bit color fluorescence image, higher values indicate greater brightness.
Addendum to:
References
- Ratcliff WC, Denison RF. Individual-level bet hedging in the bacterium Sinorhizobium meliloti. Curr Biol 2010; 20:1740 - 1744
- Ratcliff WC, Kadam SV, Denison RF. Polyhydroxybutyrate supports survival and reproduction in starving rhizobia. FEMS Microbiol Ecol 2008; 65:391 - 399
- Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 2004; 186:8172 - 8180
- Harrison JJ, Ceri H, Roper NJ, Badry EA, Sproule KM, Turner RJ. Persister cells mediate tolerance to metal oxyanions in Escherichia coli. Microbiology 2005; 151:3181 - 3195
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 2007; 5:48 - 56
- Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc Natl Acad Sci USA 2008; 105:6145 - 6149
- Vazquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol 2006; 188:3494 - 3497
- Gage DJ, Bobo T, Long SR. Use of green fluorescent protein to visualize the early events of symbiosis between rhizobium meliloti and alfalfa (Medicago sativa). J Bacteriol 1996; 178:7159 - 7166
- Gage DJ. Analysis of infection thread development using gfp- and DsRed-expressing sinorhizobium meliloti. J Bacteriol 2002; 184:7042 - 7046
- Kussell E, Kishony R, Balaban NQ, Leibler S. Bacterial persistence: A model of survival in changing environments. Genetics 2005; 169:1807 - 1814
- Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 2010; 8:1000317
- Gardner A, West SA, Griffin AS. Is bacterial persistence a social trait?. PLoS One 2007; 2:752
- Klapper I, Gilbert P, Ayati BP, Dockery J, Stewart PS. Senescence can explain microbial persistence. Microbiology 2007; 153:3623 - 3630