Abstract
Land plants and algae are now represented by about 40 genomes. Although most are incomplete, putative globins appear to be present in all the ca. 30 land plant genomes and in all except one algal genomes. The globins have either the canonical 3/3 α-helical fold characteristic of vertebrate myoglobin (Mb) or 2/2 α-helical folds, characteristic of bacterial globins with a truncated Mb-fold. In view of the fairly complete picture of the globin superfamily that is now available from analyses of over 1000 bacterial genomes and >200 other eukaryote genomes, it is now possible to seek answers to the following twin questions: what is the phylogenetic relationship of plant and algal globins to those of other eukaryotes and what is their likely bacterial origin? We summarize below the available results. Molecular phylogenetic analyses indicate that plant and algal 3/3 globins are related to bacterial flavohemoglobins and vertebrate neuroglobins. Furthermore, they also suggest that plant and algal 3/3 and group 1 2/2 Hbs originated from the horizontal gene transfers that accompanied the two generally accepted endosymbioses of a proteobacterium and a cyanobacterium with a eukaryote ancestor. In contrast, the origin of the group 2 2/2 Hbs unexpectedly appears to involve horizontal gene transfer from a bacterium ancestral to Chloroflexi, Deinococcales, Bacillli and Actinomycetes. We present additional results which indicate that the shared ancestry is likely to be with the Chloroflexi alone.
All the known plant genomes (ca. 30) contain sequences coding for globins (Hbs), which have either a 3/3 or 2/2 α-helical folds. The former comprise symbiotic Hbs, including the legHbs (Lbs) and nonsymbiotic Hbs which are divided into two classes nsHb-1 (nsHbs) and nsHb-2.Citation1–Citation3 The plant 3/3 Hbs appear to be monophyletic, with the symbiotic Hbs likely originating from the nsHb-2 globins.Citation4 The 2/2 Hbs of plants and of the related Chlorophytes and Rhodophytes (ca. 10 genomes) belong to groups 1 and 2.Citation5 The topics we would like to address are the origins of the plant and Chlorophyte Hbs.
To answer this question, we need to provide a brief overview of our current understanding of the globin superfamily phylogeny in the light of the genetic information accrued over the last 15 years. With over a 1,000 bacterial genomes now known, about two thirds contain Hbs that belong to one of three families: flavohemoglobin (F), sensor (S) and truncated Mb-fold (T).Citation6,Citation7 Although the F and S family members evince the canonical 3/3 α-helical fold characteristic of vertebrate Hb,Citation8,Citation9 the T globins have an abbreviated 2/2 α-helical fold and exist in three structural classes.Citation10,Citation11 The globin superfamily has been even more successful in eukaryotes, where apart from unicellular parasitic eukaryotes, all eukaryote groups have Hbs, ranging from >80% of the more than 100 fungal genomes to 100% in about 100 metazoan genomes. It is obvious that the present day eukaryote Hbs, including plants, must share one or more common origin with the bacterial globins. A tentative model of Hb evolution proposed that the 3/3 and 2/2 eukaryote globin genes emerged in ancestral eukaryotes via horizontal gene transfer (HGT) of bacterial F globin and T globin genes, respectively, and that the HGTs occurred via the two generally accepted endosymbiotic events involving an α-proteobacterium and a cyanobacterium that led to the formation of mitochondria and plastids, respectively.Citation12 How do the plant and Chlorophyte Hbs fit into this scenario?
We carried out molecular phylogenetic analyses of alignments of plant and algal globins with representative vertebrate globins on one hand, specifically the Ngb and Cygb families,Citation13 and with bacterial Hbs representing the F and T families.Citation5,Citation14
In the first instance, we found a close relationship between the plant 3/3 Hbs and the vertebrate Ngbs and of both to the bacterial F globins, a result compatible with the proposed model. Thus, a HGT from an ancestor of modern proteobacteria to the LECA (last eukaryote common ancestor) could represent the origin of vertebrate Ngbs and of algal and plant 3/3 Hbs. This finding is underscored by the Bayesian tree shown in , based on MUSCLE alignment of three concatenated sequences representing the various globin subfamilies (e.g., the main vertebrate groups are represented by human, bird and fish sequences). This tree also shows the expected similarity of the algal and plant group 1 and 2 T Hbs to their bacterial counterparts. Furthermore, our analysis,Citation5 also showed that the Chlorophyte group 1 T Hbs could have originated via HGT from a cyanobacterium to LECA.
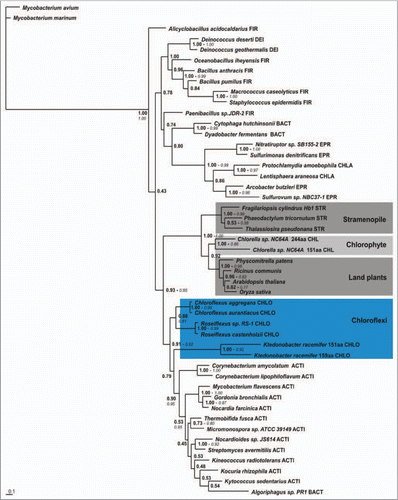
Our findings regarding the plant, Chlorophyte and Stramenopile group 2 T Hbs were quite different: their origin unexpectedly appeared to involve HGT from a bacterium ancestral to one of the following bacterial phyla, the Chloroflexi, Deinococcales, Bacillli and Actinomycetes.Citation5 Because of the size of the analyzed datasets, we were never able to obtain trees with significant branch support values. More recently we succeeded to corroborate the earlier results and refine them to show that the six known Chloroflexi group 2 T globins were indeed the closest relatives of the plant, algal and stramenopile T2 Hbs (). The phylum Chloroflexi, known formerly as “Green Non-Sulfur” bacteria, does not have a clearcut position within the bacterial phylogenetic tree.Citation15,Citation16 The recent suggestion that the root of the tree of life lies close to the Chloroflexi,Citation17 implies that the origin of algal, plant and stramenopile T2 Hbs may be quite ancient.
Abbreviations
| Cygb | = | cytoglobin |
| Hb | = | hemoglobin |
| HGT | = | horizontal gene transfer |
| F | = | flavohemoglobin |
| Mb | = | myoglobin |
| Ngb | = | neuroglobin |
| T | = | truncated myoglobin-fold globin |
Figures and Tables
Figure 1 Bayesian phylogenetic tree (Mr. Bayes 3.1.2; WAG model; 1 × 105 generations; burnin 500) based on a MUSCLE alignment of 3 concatenated sequences representing plant, Chlorophyte and Stramenopile 3/3 and 2/2 globins with 3/3 vertebrate globins and representatives of bacterial F and T families. Branch support values are bayesian posterior probabilities. FHbs, flavohemoglobins; SDFgbs, single domain F globins; Agna, Agnathan; Cygb, cytoglobin; Ngb, neuroglobin.
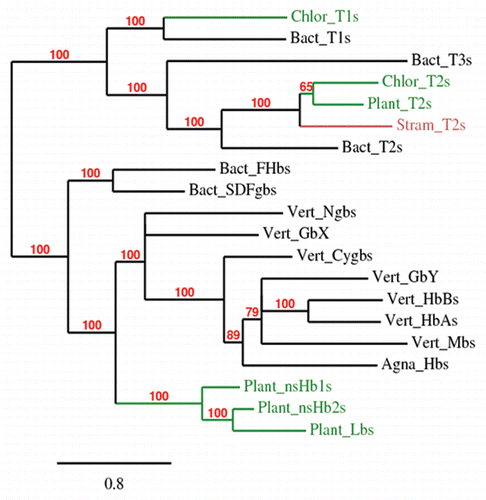
Figure 2 Bayesian phylogenetic tree (Mr. Bayes 3.1.2; WAG model; 5 × 106 generations; burnin1.5 × 106) based on concatenating MAFFT and PROBCONS alignments of representative plant, algal and stramenopile T2 globins with representative bacterial T2s and using the Mycobacterium avium and M. marinum T3 globins as outgroups. Branch support values are bayesian posterior probabilities and approximate Likelihood-Ratio Test, aLRT values (italic, if >0.5) of PhyML analysis. FIR, Firmicutes; DEI, Deinococci; BACT, Bacteroidetes; EPR, Epsilon proteobacteria; CHLA, Chlamydia; CHLO, Chloroflexi; ACTI, Actinobacteria.
Addendum to:
References
- Garrocho-Villegas V, Gopalasubramaniam SK, Arredondo-Peter R. Plant hemoglobins: what we know six decades after their discovery. Gene Funct Genom 2007; 398:78 - 85
- Hoy JA, Hargrove MS. The structure and function of plant hemoglobins. Plant Physiol Biochem 2008; 46:371 - 379
- Smagghe BJ, Hoy JA, Percifield R, Kundu S, Hargrove MS, Sarath G, et al. Correlations between oxygen affinity and sequence classifications of plant hemoglobins. Biopolymers 2009; 91:1083 - 1096
- Kakar S, Hoffman FG, Storz JF, Fabian M, Hargrove MS. Structure and reactivity of hexacoordinate hemoglobins. Biophys Chem 2010; 152:1
- Vinogradov SN, Fernández I, Hoogewijs D, Arredondo-Peter R. Phylogenetic relationships of plant and algal 3/3 and 2/2 hemoglobins to bacterial and other eukaryotic hemoglobins. Molec Plant 2010; 1:10 - 17
- Vinogradov SN, Hoogewijs D, Bailly X, Arredondo-Peter R, Guertin M, Gough J, et al. Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proc Natl Acad Sci USA 2005; 102:11385 - 11389
- Vinogradov SN, Hoogewijs D, Bailly X, Arredondo-Peter R, Gough J, Dewilde S, et al. A phylogenomic profile of globins. BMC Evol Biol 2006; 6:31
- Wu G, Wainwright LM, Poole RK. Microbial globins. Adv Microb Physiol 2003; 47:255 - 310
- Freitas TA, Saito JA, Hou S, Alam M. Globin-coupled sensors, protoglobins and the last universal common ancestor. J Inorg Biochem 2005; 99:23 - 33
- Wittenberg JB, Bolognesi M, Wittenberg BA, Guertin M. Truncated hemoglobins: a new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes and plants. J Biol Chem 2002; 277:871 - 874
- Vuletich DA, Lecomte JT. A phylogenetic and structural analysis of truncated hemoglobins. J Mol Evol 2006; 62:196 - 210
- Vinogradov SN, Hoogewijs D, Bailly X, Mizuguchi K, Dewilde S, Moens L, et al. A model of globin evolution. Gene Funct Genom 2007; 398:132 - 142
- Burmester T, Haberkamp M, Mitz S, Roesner A, Schmidt M, Ebner B, et al. Neuroglobin and cytoglobin: genes, proteins and evolution. IUBMB Life 2004; 56:703 - 707
- Fernández I, Vinogradov SN, Arredondo-Peter R. Identification and in silico characterization of a putative ancestor to land plant non-symbiotic hemoglobins from the prasinophyceae algae Micromonas and Ostreococcus. Global J Biochem 2010; 1:18 - 30
- Hanada S, Pierson BK. Dworkin M. The Family Chloroflexaceae. The Prokaryotes 2002; Springer 815 - 843
- Sutcliffe IC. A phylum level perspective on bacterial cell wall architecture. Trends Microbiol 2010; 18:464 - 470
- Valas RE, Bourne PE. Structural analysis of polarizing indels: an emerging consensus on the root of the tree of life. Biol Direct 2009; 4:30