Abstract
The divergent eukaryotic unicellular organism Giardia intestinalis is an intestinal parasite in humans and various animals. An analysis of a draft genome sequence suggested that G. intestinalis has a much simpler genome organization and gene repertoire than those of other model eukaryotic organisms (e.g., Arabidopsis and human). This general picture of the G. intestinalis genome seemingly agrees with the fact that only four spliceosomal (cis-spliced) introns have been identified in this organism to date. We have recently shown that G. intestinalis possesses a unique gene expression system incorporating spliceosome-mediated trans-splicing. Some protein-coding genes in G. intestinalis are split into multiple pieces in the genome and each gene fragment is independently transcribed. Two particular pre-mRNAs directly interact with each other by forming an intermolecular-stem structure and are then trans-spliced into a mature mRNA by spliceosomes. We believe that this trans-splicing secondarily arose from the system that excises canonical (cis-splicing) introns. Based on these findings, we suspect that similar phenomena—split genes and post-transcriptional assemblage of their transcripts via trans-splicing—may be prevalent in more distinct eukaryotic lineages than previously known, particularly in organisms possessing ‘intron-poor’ genomes.
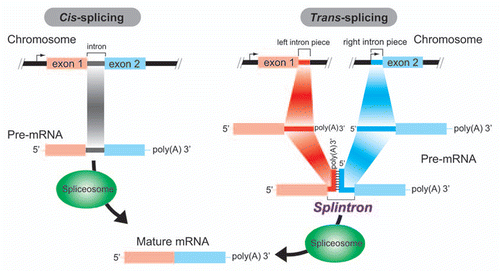
In eukaryotic genes, a coding region is often interrupted by non-coding sequence stretches called introns. As premature messenger RNAs (pre-mRNA) including introns are initially generated by transcription, the elimination of introns and concurrent joining of coding regions (exons) by splicing is required to produce mature translatable mRNAs (, left). In eukaryotic systems, intron splicing is mediated by a large molecular machines called spliceosomes composed of ribonucleoprotein particles and many other components.Citation1 Introns excised by spliceosomes (so-called spliceosomal introns) are exclusively found in eukaryotes, and thus spliceosomal introns are hallmarks of eukaryotic nuclear genome organization. Nevertheless, some eukaryotes possess very few spliceosomal introns in their genomes as compared to well known model organisms (e.g., humans and Arabidopsis).Citation2 The divergent eukaryotic intestinal protistan parasite Giardia intestinalis is one of such intron-poor species.Citation3 In the Giardia draft genome, the genes encoding many spliceosomal components have been identified, but only four spliceosomal introns have been reported so far.Citation3,Citation4 Thus, Giardia, as well as other intron-poor organisms, is a key organism to understand the evolutionary dynamics of spliceosomal introns and spliceosomes in eukaryotes.
Recently, we reported a novel spliceosome-mediated trans-splicing system in Giardia.Citation5 In the Giardia genome, two distant loci that are transcribed as two distinct polyA+ pre-mRNAs encode the N- and C-terminal amino acid sequences of HSP90. More interestingly, the non-coding regions of these pre-mRNAs possess stretches of nucleotides that are complementary to each other and thus have the potential to form a 26 bp-long intermolecular stem structure (, right). The putative stem structure is flanked by the sequence motifs conserved amongst the four cis-spliced spliceosomal introns previously identified in Giardia. We experimentally showed that the two pre-mRNAs are efficiently processed into a single, mature mRNA by spliceosomes, which recognize the intron-like motifs in the two separate RNA molecules. The trans-splicing of the Giardia hsp90 gene was also reported by another research group, and they confirmed that no translation of the pre-mRNAs occurs.Citation6 Surprisingly, we found that the same trans-splicing system is involved in the gene expression of C-terminal outer arm dynein beta chain (OADβ) in Giardia.Citation5 Henceforth here, we designate these split spliceosomal introns as “splintrons.”
Splintrons are important for understanding the post-transcriptional RNA processing machinery of Giardia. Additional splintrons likely remain undetected in the Giardia genome, because an extensive in silico survey explicitly aiming to detect splintrons has yet to be done. Nevertheless, our estimated ratio of splintrons to canonical introns in the Giardia genome has implications for the major functions of the Giardia spliceosomes. Previously-conducted surveys identified only four cis-spliced (canonical) introns in total whereas we have identified three splintrons (one in hsp90 and two in OADβ genes).Citation3,Citation5 Therefore it seems that Giardia possesses similar numbers of splintrons versus canonical introns in its genome. We expect that Giardia spliceosomes likely contain non-canonical spliceosomal subunits exclusively dedicated to trans- versus cis-splicing, as observed in trypanosomatid and nematode spliceosomes.Citation7 Thus the gene expression system in Giardia may turn out to be more complex than we currently think.
Are splintrons unique to this Giardia intestinalis genome? The answer is “no.” We identified the splintron in hsp90 and OADβ genes in phylogenetically diverged Giardia isolates,Citation5 and the nematode Caenorhabditis elegans has been also known to possess a single splintron amongst ∼19,000 intron-containing genes.Citation8,Citation9 Although this single case indicates a very minor role for splintron transsplicing in the gene expression system in C. elegans, the splintrons in C. elegans and Giardia are strikingly similar in that they form intermolecular stem structures between the split intron pieces (i.e., non-coding regions in two pre-mRNAs). It is very surprising that such an extremely distinct trans-splicing system has independently emerged on the branches leading to C. elegans and Giardia. Perhaps we should be searching for splintrons in other newly-characterized eukaryotic genomes, particularly in those that are “intron-poor” as Blumenthal has recently suggested.Citation10
Here, as well as in our original article, we stress the significance of splintrons for understanding the evolution of spliceosomal introns. We argue that splintrons are unlikely to represent the ancestral form of spliceosomal introns. The latter in eukaryotic (nuclear) genomes were most likely derived from group II intronsCitation11 that were introduced into eukaryotes with the mitochondrial symbiont.Citation12 The vast majority of group II introns are cis-spliced and it seems likely that they evolved directly into the cis-spliced spliceosomal introns in the nucleus of a common ancestor of all extant eukaryotes. In this view, all trans-splicing cases including splintrons in Giardia and C. elegans are secondarily derived from a cis-splicing ancestor.
Finally, although many have suggested that Giardia is a “deep-branching” eukaryotic lineage, it is widely thought that this position is due to the long-branch attraction artefact in phylogenetic tree reconstruction.Citation13,Citation14 Therefore we should be cautious to treat the significance of Giardia splintrons to intron evolution separately from the “deep-branching” status of Giardia inferred by potentially biased analyses.
Abbreviations
| HSP90 | = | heat shock protein 90 |
| OAD-β | = | outer arm dynein beta chain |
| pre-mRNA | = | preliminary mRNA |
Figures and Tables
Figure 1 Depiction of intron splicing processes in Giardia. Left: Cis-splicing of a canonical spliceosomal intron. A single, continuous pre-mature mRNA (pre-mRNA) including introns is transcribed from a single locus in a genome. An arrow indicates the transcription initiation site at the 5′ upstream region of exon 1. Canonical introns are then cis-spliced by spliceosomes. Right: Trans-splicing of a splintron. Two poly-A+ pre-mRN As including a “left splintron piece” and a “right splintron piece” are independently transcribed from two distant loci in the genome (two different transcription initiation sites are highlighted by arrows). As the pre-mRNA forms an intermolecular stem structure, the pre-mRNA complex can be recognized as the substrate for spliceosomes.
Acknowledgments
R.K. was a research fellow supported by JSPS for Young Scientists (210528). This work was supported by a part of a JSPS grant awarded to Y.I. (21370031) and T.H. (20570219) and a CIHR MOP-62809 grant awarded to A.J.R.
Addendum to:
References
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 2003; 12:5 - 14
- Keeling PJ, Slamovits CH. Causes and effects of nuclear genome reduction. Curr Opin Genet Dev 2005; 15:601 - 608
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 2007; 317:1921 - 1926
- Nixon JE, Wang A, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, et al. A spliceosomal intron in Giardia lamblia. Proc Natl Acad Sci USA 2002; 99:3359 - 3361
- Kamikawa R, Inagaki Y, Tokoro M, Roger AJ, Hashimoto T. Split introns in the genome of Giardia intestinalis are excised by spliceosome-mediated trans-splicing. Curr Biol 2011; 21:311 - 315
- Nageshan RK, Roy N, Hehl AB, Tatu U. Post-transcriptional repair of a split heat shock protein 90 gene by mRNA trans-splicing. J Biol Chem 2011; 286:7116 - 7122
- Günzl A. The pre-mRNA splicing machinery of trypanosomes: Complex or simplified?. Eukaryot Cell 2010; 9:1159 - 1170
- Fischer SEJ, Butler MD, Pan Q, Ruvkun G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 2008; 455:491 - 496
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 1998; 282:2012 - 2018
- Blumenthal T. Split genes: another surprise from Giardia. Curr Biol 2011; 21:162 - 163
- Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet 2004; 38:1 - 35
- Roger AJ, Doolittle WF. Why introns-in-pieces?. Nature 2000; 364:289 - 290
- Philippe H, Philippe L, Brinkmann H, Budin K, Germot A, Laurent J, et al. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc R Soc Land B 2000; 267:1213 - 1221
- Roger AJ, Hug LA. The origin and diversification of eukaryotes: problems with molecular phylogenetics and molecular clock estimation. Phil Trans R Soc B 2006; 361:1039 - 1054