?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
There are three sources of free energy for cells: chemical potential, electrical potential and mechanical potential. There is little known about the last one since there have not been simple ways to measure stress in proteins in cells. We have now developed genetically encoded force sensors to assess the stress in fibrous proteins in living cells. These FRET based fluorescence sensors can be read out at video rates and provide real time maps of the stress distribution in cells, tissues and animals. The sensors can be inserted into specific proteins and in general do not disturb the normal function or anatomy. The original sensors used mutant GFPs linked by elastic linkers. These sensors provide a linear output with applied stress but the response is linear in strain. To improve contrast and dynamic range we have now developed a new class of sensors that are smaller making them less invasive, and have much higher intrinsic sensitivity since force modulates the angle between the donor and acceptor much more than the distance between them. Known as cpstFRET, the probe shows improved biocompatibility, wider dynamic range and higher sensitivity.
Mechanical Force and Biology
All cells are mechanosensitive. This spans single cell bacteria and amoebas, fungi, curling tendrils and twirling vines, from insects to cold blooded reptiles to mammals. Biological systems all utilize three sources of free energy: chemical potential, electrical potential and mechanical potential. The flow of energy between these sources is linked by numerous feedback loops such as stretch-sensitive channels,Citation1,Citation2 but little is known of the mechanical potential since there have been no simple ways to measure it. We know that mechanical force triggers prompt electrical responses that create the senses of touch, hearing, local gravity and the input needed for coordination of the voluntary musculature. Physical forces alter the structure of the cytoskeleton, reorient nuclei and redistribute organelles that in turn are coupled to morphology, migration and sensitivity to other external stimuli.Citation3–Citation5 All proteins are mechanosensitive since they are elastic—these properties are at the heart of molecular dynamic computations.Citation6–Citation8 Numerous studies have also indicated that mechanical forces can regulate gene expression.Citation9–Citation12 There are feedback loops incorporating mechanical energy for a vast array of cell activities such as embryonic development and differentiation,Citation13,Citation14 meiosis and mitosis,Citation15,Citation16 cancer malignancy and metastasis,Citation17,Citation18 blood vessel angiogenesis,Citation19–Citation22 bone growth and osteoporosis and lung, heart and skeleton muscle metabolism.Citation23
Techniques for Studying Cell Mechanics to Date
As a universal component of biological systems factor, mechanical forces need to be measured in living cells. Most of the work on cell mechanics has been devoted to applying external perturbations and recording various responses, but the cytoskeleton (and the extracellular matrix or ECM) are the components of the cell that are responsible for reacting to external stress. Both of them are heterogeneous and anisotropic arrays of fibrous proteins cross linked with stress sensitive bonds so that there should be no universal response from all proteins. We recently created molecular force sensors that can be genetically encoded and inserted in the middle of fibrous proteins to measure the stress in specific proteins in living cells in real time. Before we discuss the details of how to measure protein stress we will discuss how forces are applied to cells and how the mean stresses are measured.
There are five major techniques used to study cell mechanics: elastic substrates,Citation24 microfabricated cantilevers and surfaces,Citation25,Citation26 laser tweezers,Citation27 atomic force microscopyCitation28 and microfabricated elastic probes.Citation29 Elastic substrates detect mechanical forces exerted by the cell by mapping the strain distribution in the substrate using fluorescent probes,Citation30 the formation of wrinkles in the surfaceCitation31 or imaging of other optical markers. From the measured strain and knowledge of elastic properties of the substrate, the stress distribution can be mapped and calibrated. For non muscle cells in culture, and the peak forces are in the range of nanoNewtons.Citation31 Laser tweezers apply calibrated forces to micron sized beads bund to cells and these measurements are mostly confined to the extracellular surface where the beads can be readily applied. The applied forces are in the range of 0.1–300 pN.Citation32 Atomic force microscopes apply forces through submicron dimension micro-fabricated probes and the method can be used for single molecule measurements in the pN range to whole cell measurement in the nN range. Like the laser tweezers, the AFM can only be applied to the extracellular surface of cells,Citation33–Citation35 but the technique can measure the compliance of a small region of a cell. The soft elastic probes such as those made with PDMS micro-needles that measurably deform when pulled on by a cell.Citation36 None of these techniques can be use to examine intracellular stresses without disrupting the cells. Nor are they easily applicable to stimulation of tissues or in situ in living animals. Thus, to explore the distribution of stress inside cells we need to have stress probes that can be used non-invasively in cells and can probe the different proteins that make up the cytoskeleton and ECM.
Transforming Mechanical Forces into Optical Signals
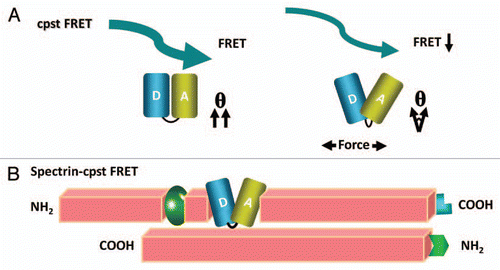
To study forces in specific proteins, sensors need to be integrated into host proteins hence all the published sensors are genetically encoded. The first such force sensor was named “stretch sensitive FRET” (stFRET) and is shown in ().Citation37 It consisted of mutant GFPs called CeruleanCitation38 and VenusCitation39 linked by a stable α-helix.Citation40 We picked this linker because the helix with a length (5 nm) equal to the characteristic distance (Ro) for energy transfer for those fluorophores and it is stable. Equation 1 shows that when R, the distance between donor and acceptor, is equal to Ro there is 50% energy transfer:
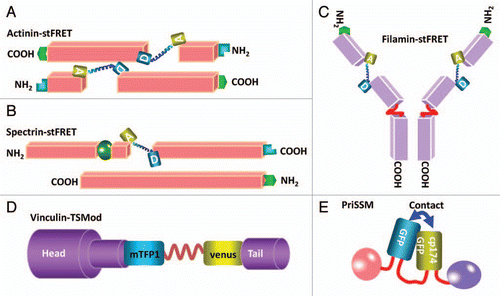
The strain sensitivity, the derivative of E with respect to R, is maximal at R = R0. In the host protein, stress creates strain in the probe linker and changes R, changing the FRET efficiency. We genetically integrated stFRET into α-actinin, filamin, non-erythroid spectrin, collagen-19 and non-muscle myosin 2A and 2BCitation37,Citation41,Citation42 (). Targeting the sensor near the middle of the proteins and avoiding known functional domains usually produces normal proteins. Only in the case of myosins have we not yet found a non-perturbing site. The general non-perturbing nature of the probe was emphasized when we created transgenic Caenorhabditis elegans with mutated collagen-19-stFRET.Citation43 We made three mutants with stFRET inserted at different locations, and the most central location yielded normal anatomy and animal behavior. We have now created transgenic mice with actinin probes and the animals create pre-labeled cells for studies of cell mechanics in vitro, in vivo and in situ.
Probe linkers could introduce dynamic errors into protein function. There is a general question of whether the probe should be stiffer of softer than the host protein. If it is stiffer, the probe termini will move less under stress. If it is softer, the probe termini will move more and there will be less stress at the same strain. When the probe is stiff the force sensitivity is reduced and when it is more compliant there is more sensitivity. There is clearly no universal optimum, but the most efficient coupling of mechanical energy from the host to the probe occurs when the compliance of the two are equal. As a test we made the linker out of a spectrin repeat since a number of our hosts utilize these repeats and we called the new construct sstFRET ().Citation42 Spectrin repeats domains are composed of three folded β-helices and are conserved in spectrin superfamily proteins including β-actinin, dystrophin, utrophin and kalirin.Citation44 These proteins cross link actin fibers and/or directly connect fibers to the cell membrane. We found that using the repeat domain as a linker provided higher compliance than the helix of stFRET. We tested actinin-sstFRET in human embryonic kidney cell (HEK) and bovine aortic endothelial cells (BAEC). The actinin distributions and functions were normal, as defined by comparison to terminal GFP tagged version of the same protein. We found significant constitutive stress and force modulation during cell contraction, extension and migration.Citation42 In both actinin and filamin in migrating cells we found similar stress patterns with stFRET and sstFRET.
We created a transgenic mouse line with actinin-sstFRET by randomly incorporating the construct into genome using the pronucleus method. PCR confirmed the presence of the correct genes in adult mice. The mice have now been bred to F3 without showing morphological abnormality or abnormal behavior, but we observed little fluorescence in the F3 mice and are establishing whether this is an expression problem. We are creating new strains using other protein hosts.
With time, other research groups began using similar force sensors. Grashoff et al. created a variant called the “tension sensor module” (TSMod) consisting of the FRET pair Venus and mTFP1,Citation45 connected by a 40 amino acid elastic domain derived from silk protein flagelliform ().Citation46 TSMod was incorporated into the flexible linker domain between head and tail domains of vinculin (). They estimated the tension applied to focal adhesions through vinculin. As we found with actinin and other cytoskeletal proteins, vinculin is under high stress at the leading edge of migrating cells and lower stress at the lagging edge. The three sensors discussed above are all FRET based and increased stress leads to decreased FRET. Uyeda's group created another sensor based on proximity imaging (PRIM), named PRIM-based strain sensor module (PriSSM) ().Citation47 PriSSM is a heterodimer formed by wild type GFP and circularly permutated GFP (cp174GFP) with linker domain AS(GGS)9 between them (). PRIM depends on direct contact between 2 GFP molecules, interactions that can lead to structural perturbations and spectral changes.Citation48 The authors flanked the sensor with myosin 2 motor domains and directly visualized the stress in myosin when it interacts with F-actin ().Citation47 For all these sensors, the force sensitivity is estimated to be in the range of 1–10 pN. We measured our sensitivity in solution by stretching the probe with “DNA springs.” Following the work of Zocchi's lab,Citation7 we attached a 60mer of single stranded DNA between the donor and acceptor.Citation42 Being floppy (having a short persistence length), ssDNA does not exert much force on the probe. However, when the complementary strand of DNA is added, the resulting dsDNA is much stiffer and pushes the fluorophores apart with a force of 5–7 pN.Citation49 We found that these forces extended our linker, significantly reducing FRET. Another group measured stiffness of the linker alone using laser tweezersCitation46 and found similar force sensitivity. Stress of the linker plays the predominant role in the compliance since the fluorophores can withstand more than 100 pN without unfolding.Citation50
How sensitive can we make a probe? Probe sensitivity refers to the force induced change in mean signal relative to background noise. The signal comes from changes in R under stress and this is a function of probe compliance. The more compliant the probe, the larger the change in R with a given stress, so why not use the most compliant linker? There are three reasons. Highly compliant probes decrease the stress in the host possibly disturbing normal function of the protein. Secondly, more compliant probes tend to be larger and thus create larger perturbation of the host. And finally the most fundamental factor is that more compliant the linker, the more thermal vibrations will wiggle the donor an acceptor and increase the background noise. You don't gain much sensitivity by increasing the compliance. However, increasing quantum yield and transfer efficiency improves the signal level and hence the signal to noise ratio by decreasing photon arrival noise. The signal to noise ratio is also determined by the contrast created by the probe, i.e., the dynamic range. In the case of the stretched linker, when R = R0 the FRET efficiency is a linear function of strain so that one cannot obtain a large dynamic range. But by letting force vary the angle between the donor and acceptor, the potential contrast can approach 100%.
A FRET Based Sensor Using Angular Sensitivity
In an attempt to expand the dynamic range and sensitivity of the sensor and to reduce its size we developed a new type of sensor that senses the angular displacement. This relative orientation of the donor and acceptor dipoles has a strong angular dependence of the FRET efficiency summarized in the term commonly called κ2. Parallel dipoles have κ2= 4 and perpendicular dipoles have κ2= 0. We demonstrated the angular dependence of the efficiency in our original workCitation37 by sequentially deleting or adding amino acids to the helical linker.Citation37 Each amino acid deletion or addition rotated the angle of the termini by 100 degrees and change the distance a relatively small amount, but deleting an entire turn changed the distance by a large amount and left the angle unchanged. By fitting the observed FRET efficiencies to a set of simultaneous trigonometric equations involving R and the various angles we derived the angular sensitivity and the geometry of the fluorophores. That study predicted that if we can arrange the donor and acceptor to be adjacent and parallel at rest, torsion forces exerted on the fluorophores by the host protein will twist them apart to higher angles leading to dramatic decreases in FRET, i.e., high contrast.
This new probe is called cpstFRET, since we circularly permutated the donor and acceptor to form cpCerulean and cpVenus (paper submitted). We linked them tightly together at one end with a flexible linker and in the resulting probe the fluorophores are nearly parallel at rest generating robust FRET with an efficiency E = 75%. When we stretched the dimer with a DNA spring, cpstFRET showed a significant decrease in FRET demonstrating a much wider dynamic range than previous sensors. Furthermore, the small linker reduced the net probe size by ∼30%, from 74 kDa to 54 kDa (). We incorporated cpstFRET into spectrin in HEK293 cells and bovine aortic endothelial cells (BAECs) and examined the stress in spectrin at rest and during cell migration (). We found that there is a rich landscape of internal forces and that these cannot be inferred from the cell anatomy. These sorts of probes are opening up cellular biomechanics to new insights.
Outlook
Cell mechanics undergo dynamic changes during normal growth and development and these forces are now open to study, a largely unexplored domain. For example, we used the probes to demonstrate that in hypotonic swelling, osmotic stresses are predominately in the cytoskeleton and not the cortex. Thus animal cells behaves more like sponges than the familiar model of semipermeable bags.Citation33 With transgenic animals that supply built in probes, it is now possible to study forces in not only cells, but tissues and whole animals in real time, subject only to the capabilities of FRET imaging. We envisage recording the stress in cytoskeletal proteins in the heart during a normal and arrhythmic cardiac cycles, stress in the voluntary musculature and connective tissue of swimming zebra fish, measuring the shear stress in the glycocalyx during blood flow and so on. Fun times ahead.
Figures and Tables
Figure 1 Cartoon diagrams of genetically encoded force sensors where the letter A in the fluorophore represents the acceptor Venus, the letter D represents the donor Cerulean. The arrows represent energy transfer from donor to acceptor. (A) stFRET; (B) sstFRET. (C) TSMod, mTFP1 is the donor and Venus is the acceptor. (D) PriSSM, using wild-type GFP and circularly permutated GFP for PRIM dimers.
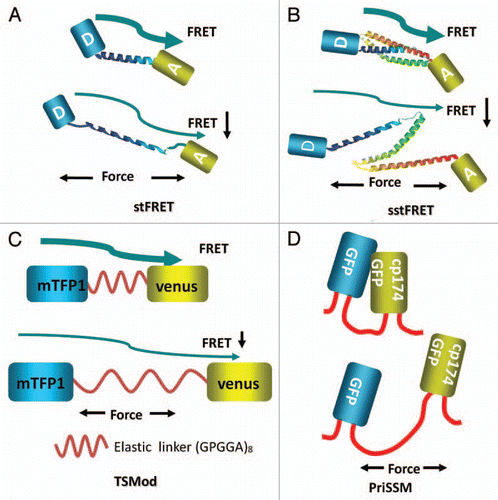
Figure 2 Diagrams of inserting the force probes into host proteins. (A) Actinin-stFRET forming anti-parallel dimers. (B) Spectrin-stFRET exists as a hetero-dimer with α and β subunits. (C) Vinculin-TSMod associating at focal adhesions as monomers. (D) A protein construct of PriSSM with myosin motors on the ends.
Acknowledgements
This work was supported by a grant from US National Institutes of Health and The Children's Fund of Buffalo.
References
- Martinac B. Mechanosensitive ion channels: molecules of mechanotransduction. J Cell Sci 2004; 117:2449 - 2460
- Sachs F. Stretch-activated ion channels: what are they?. Physiology (Bethesda) 2010; 25:50 - 56
- Chen CS, Tan J, Tien J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu Rev Biomed Eng 2004; 6:275 - 302
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA 1997; 94:849 - 854
- Entcheva E, Bien H. Mechanical and spatial determinants of cytoskeletal geodesic dome formation in cardiac fibroblasts. Integr Biol (Camb) 2009; 1:212 - 219
- Moskowitz DW. Is “somatic” angiotensin I-converting enzyme a mechanosensor?. Diabetes Technol Ther 2002; 4:841 - 858
- Zocchi G. Controlling proteins through molecular springs. Annu Rev Biophys 2009; 38:75 - 88
- Saghatelian A, Guckian KM, Thayer DA, Ghadiri MR. DNA detection and signal amplification via an engineered allosteric enzyme. J Am Chem Soc 2003; 125:344 - 345
- Na S, et al. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci USA 2008; 105:6626 - 6631
- Song G, Luo Q, Xu B, Ju Y. Mechanical stretch-induced changes in cell morphology and mRNA expression of tendon/ligament-associated genes in rat bone-marrow mesenchymal stem cells. Mol Cell Biomech 2010; 7:165 - 174
- Chan MW, Chaudary F, Lee W, Copeland JW, McCulloch CA. Force-induced myofibroblast differentiation through collagen receptors is dependent on mammalian diaphanous (mDia). J Biol Chem 2010; 285:9273 - 9281
- Zhang F, et al. Compressive force stimulates the gene expression of IL-17s and their receptors in MC3T3-E1 cells. Connect Tissue Res 2010; 51:359 - 369
- Adamo L, Naveiras O, Wenzel PL, McKinney-Freeman S, Mack PJ, Gracia-Sancho J, et al. Biomechanical forces promote embryonic haematopoiesis. Nature 2009; 459:1131 - 1135
- North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, et al. Hematopoietic stem cell development is dependent on blood flow. Cell 2009; 137:736 - 748
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 2008; 133:1188 - 1201
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol 2009; 19:749 - 761
- Kumar S, Weaver VM. Mechanics, malignancy and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev 2009; 28:113 - 127
- Plodinec M, Schoenenberger CA. Spatial organization acts on cell signaling: how physical force contributes to the development of cancer. Breast Cancer Res 2010; 12:308
- Cunningham KS, Gotlieb AI. The role of shear stress in the pathogenesis of atherosclerosis. Lab Invest 2005; 85:9 - 23
- Egginton S. Angiogenesis—may the force be with you!. J Physiol 2010; 588:4615 - 4616
- Ino K, Okochi M, Honda H. Application of magnetic force-based cell patterning for controlling cell-cell interactions in angiogenesis. Biotechnol Bioeng 2009; 102:882 - 890
- Siasos G, Tousoulis D, Siasou Z, Stefanadis C, Papavassiliou AG. Shear stress, protein kinases and atherosclerosis. Curr Med Chem 2007; 14:1567 - 1572
- Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev Cell 2006; 10:11 - 20
- Munevar S, Wang Y, Dembo M. Traction force micros-copy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J 2001; 80:1744 - 1757
- Galbraith CG, Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci USA 1997; 94:9114 - 9118
- Saif MT, Sager CR, Coyer S. Functionalized biomicro-electromechanical systems sensors for force response study at local adhesion sites of single living cells on substrates. Ann Biomed Eng 2003; 31:950 - 961
- Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 2003; 424:334 - 337
- Kirmizis D, Logothetidis S. Atomic force microscopy probing in the measurement of cell mechanics. Int J Nanomedicine 2010; 5:137 - 145
- Leporatti S, Sczech R, Riegler H, Bruzzano S, Storsberg J, Loth F, et al. Interaction forces between cellulose microspheres and ultrathin cellulose films monitored by colloidal probe microscopy-effect of wet strength agents. J Colloid Interface Sci 2005; 281:101 - 111
- Mizutani T, Haga H, Kawabata K. Cellular stiffness response to external deformation: tensional homeostasis in a single fibroblast. Cell Motil Cytoskeleton 2004; 59:242 - 248
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol 2001; 3:466 - 472
- Nieminen TA, Knoner G, Heckenberg NR, Rubinsztein-Dunlop H. Physics of optical tweezers. Methods Cell Biol 2007; 82:207 - 236
- Spagnoli C, Beyder A, Besch S, Sachs F. Atomic force microscopy analysis of cell volume regulation. Phys Rev E Stat Nonlin Soft Matter Phys 2008; 78:31916
- Beyder A, Sachsb F. Electromechanical coupling in the membranes of Shaker-transfected HEK cells. Proceedings of the National Academy of Sciences of the United States of America 2009; 106:6626 - 6631
- Besch S, Snyder KV, Zhang RC, Sachs F. Adapting the Quesant (c) Nomad (TM) atomic force microscope for biology and patch-clamp atomic force microscopy. Cell Biochemistry and Biophysics 2003; 39:195 - 210
- du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, et al. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA 2005; 102:2390 - 2395
- Meng F, Suchyna TM, Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J 2008; 275:3072 - 3087
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 2004; 22:445 - 449
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 2002; 20:87 - 90
- Chen C, Brock R, Luh F, Chou PJ, Larrick JW, Huang RF, et al. The solution structure of the active domain of CAP18—a lipopolysaccharide binding protein from rabbit leukocytes. FEBS Lett 1995; 370:46 - 52
- Meng F, Sachs F. stFRET, a Novel Tool to Study Molecular Force in Living Cells and Animals 2008; 14 Proquest Dissertation 1757060081
- Meng F, Sachs F. Visualizing dynamic cytoplasmic forces with a compliance-matched FRET sensor. J Cell Sci 2011; 124:261 - 269
- FMeng F, Suchyna TM, Lazakovitch E, Gronostajski RM, Sachs F. Real Time FRET Based Detection of Mechanical Stress in Cytoskeletal and Extracellular Matrix Proteins. Cell Mol Bioeng 2011; 4:148 - 159
- Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett 2002; 513:119 - 123
- Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J 2006; 400:531 - 540
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 2010; 466:263 - 266
- Iwai S, Uyeda TQ. Visualizing myosin-actin interaction with a genetically-encoded fluorescent strain sensor. Proc Natl Acad Sci USA 2008; 105:16882 - 16887
- De Angelis DA, Miesenbock G, Zemelman BV, Rothman JE. PRIM: proximity imaging of green fluorescent protein-tagged polypeptides. Proc Natl Acad Sci USA 1998; 95:12312 - 12316
- Tseng CY, Wang A, Zocchi G, Rolih B, Levine AJ. Elastic energy of protein-DNA chimeras. Phys Rev E Stat Nonlin Soft Matter Phys 2009; 80:61912
- Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc Natl Acad Sci USA 2004; 101:16192 - 16197