Abstract
Messenger RNA (mRNA) localization plays an important role in various cellular functions. To date, two general mechanisms have been identified for intracellular mRNA localization. The first one was identified by Blobel and colleagues more than three decades ago, by which mRNAs encoding for membrane and secreted proteins are targeted to the endoplasmic reticulum (ER) in a signal peptide dependent manner.1 The second mechanism is for the intracellular targeting of mRNAs encoding cytosolic proteins, which is dependent on specific sequence on the mRNA called zipcode.2 Recently, we have identified a new mechanism which targets Dia1 mRNA to the perinuclear ER in a zipcode-independent manner, even though the mRNA encodes a cytosolic protein.3 Here, we provide an updated discussion on how the Dia1 mRNA is targeted and what might be its physiological significance.
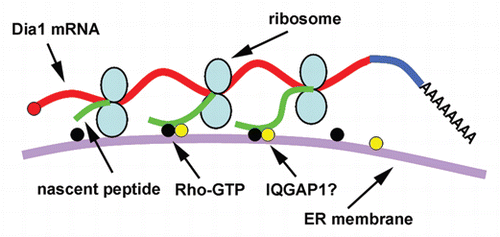
In one of our recent reports, we demonstrate that Dia1 mRNA is targeted to the perinuclear ER by a new mechanism that is independent of an RNA zipcode.Citation3 Instead, the Dia1 mRNA targeting requires active translation and interactions between Dia1 nascent peptide with active Rho, providing the first evidence for a direct role of Rho in mRNA localization and indicating a role of nascent peptide in guiding the intracellular localization of mRNAs encoding cytosolic proteins. Because of limit of space in that article, we only discussed some of the most likely scenarios as how Dia1 mRNA might be localized on the ER. The following is an update of our discussion as to how and why Dia1 mRNA is localized.
Does IQGAP1 Play a Role in Dia1 mRNA Localization?
We have demonstrated that localization of Dia1 mRNA to the perinuclear ER requires a Dia1 nascent peptide which must contain both the Rho GTPase binding domain (GBD) and diaphanous inhibitory domain (DID).Citation3 While it is clear that GBD is for binding to Rho-GTP, the role of DID in Dia1 mRNA localization is less clear. In our previous working model we speculated that DID might be required to enhance or stabilize the interactions between the GBD of Dia1 nascent peptide and Rho-GTP given that DID can physically contact with Rho-GTP in a crystal structure.Citation4,Citation5 However, it is equally possible that the DID is required for binding to another protein on the ER. It is therefore interesting to notice that Brandt and Grosse reported that IQGAP1 interacts with the DID of Dia1.Citation6 IQGAP1 is a scaffold protein with multiple functions, which localizes to the perinuclear region and leading edge in fibroblasts.Citation7,Citation8 Its binding to DID of Dia1 requires Rho binding to the GBD of Dia1. Given that the GBD of Dia1 nascent peptide binds to Rho-GTP,Citation3 this will permit the interactions between IQGAP1 and the DID. Hence, interactions between IQGAP1 and the DID of Dia1 nascent peptide is expected to enhance the anchorage of the ribosome-mRNA-nascent peptide complex on the ER.
Prompt Initiation of Translation and Pausing of Translational Elongation
When exogenous Dia1 was expressed in transfected fibroblasts, it was often observed that the ectopically expressed Dia1 mRNA was enriched in a narrow zone surrounding the nucleus.Citation3 We predict that this is due to a prompt translation of the Dia1 mRNA once it exits the nuclear pore as translation is necessary for the Dia1 mRNA localization. It is unlikely that such pattern of mRNA localization could be resulted from a pool of Dia1 mRNA which is first diffused and translated in the cytoplasm then is transported to the perinuclear ER. This argument is based on that if Dia1 mRNA is first translated in the cytoplasm, the relatively enriched Rho-GTP in the cytoplasm and plasma membrane may keep the mRNA there because interactions between Dia1 nascent peptide and Rho-GTP dictates Dia1 mRNA localization. A test for this question would be to determine whether delocalized Dia1 mRNA can localize to the perinuclear ER by first delocalizing the Dia1 mRNA using puromycin and then washing-off the drug (in the presence of transcription inhibitor to prevent the accumulation of newly transcribed Dia1 mRNA). In addition to prompt translation, pausing during translational elongation may play a role in Dia1 mRNA localization. It is known that translation of mRNAs encoding membrane and secreted proteins pauses after the signal peptide is synthesized.Citation9 This pausing is important for the subsequent co-translational localization of these mRNAs on the ER. In another case, a nascent peptide motif pauses the translation of XBP1u mRNA for the splicing of an intron on the ER.Citation10 It is generally believed that translation pausing helps co-translational folding of nascent peptides and even interactions of the folded domain with binding partners for regulatory purposes.Citation11,Citation12 Our model predicts that a Dia1-mRNA-ribosome complex is tethered on the ER through Dia1 nascent peptides.Citation3 Therefore multiple nascent-peptide-ribosome complexes on each Dia1 mRNA are required to ensure that at any given moment there is at least one nascent peptide which is bridging the ribosome-mRNA and the ER (also see ). It is feasible that translation pausing of Dia1 mRNA will increase the number of the ribosome on each mRNA. In addition, the pausing may provide more time for the folding of Dia1 nascent peptide hence facilitating its interactions with Rho-GTP. In this regard, it is interesting to notice that there is some similarity between a short sequence in the GBD of Dia1 and the translation pausing motif of XBP1uCitation10 (). Given that the GBD-containing N-terminal fragment of Dia1, which is predicted to have much fewer number of ribosome on each mRNA due to its much shorter length, is sufficient for its mRNA localization,Citation3 this further raises the possibility that translation pausing plays a role in Dia1 mRNA localization.
The Physiological Significance of Dia1 mRNA Localization
The perinuclear ER localization of Dia1 mRNA was a surprise to us given that no role has been proposed for Dia1 protein on the ER. Rather, Dia1 has been proposed to work in other compartments such as the leading edge.Citation6 Then why do the cells localize the Dia1 mRNA and synthesize Dia1 protein on the ER instead of localizing the Dia1 mRNA to its site of function at the leading edge as in the case for the Arp2/3 mRNAs?Citation13,Citation14 Although there is currently no clear answer to this question, our preliminary data do suggest that the perinuclear ER localization of Dia1 mRNA may be important for cellular function as de-localization of Dia1 mRNA in fibroblasts could impair cell migration (Liao G and Liu G, unpublished). Because one of the functions of mRNA localization is to avoid inappropriate protein interactions, it is possible that local synthesis of Dia1 on the ER is to prevent its premature interactions with other proteins until it is functionally ready (e.g., folded and formation of dimer). Another possible reason for the local synthesis of Dia1 on the ER is its potential involvement in ER expansion. It is known that Dia1 regulates microtubules (MT) stability and MT regulate ER extension.Citation15,Citation16 Therefore, the first job of the newly synthesized Dia1 might be involved in the interactions with MT in the perinuclear region. Finally, because IQGAP1 is required for the recruitment of Dia1 to the leading edge of fibroblasts,Citation6 the above mentioned local interactions between the DID of Dia1 nascent peptide and IQGAP1 on the ER may precondition IQGAP1 for this purpose as the nascent Dia1 peptides are in a Rho-GTP bound state which is a prerequisite for the DIDIQGAP1 interactions.
Figures and Tables
Figure 2 Predicted potential translation pausing motif (CTR) in chicken and human Dia1 protein sequences. The CTR sequences are from reference Citation10. The CTR in both chicken and human Dia1 are overlapped with a portion of the GBD.

Acknowledgments
This work is supported by NIH grant R01GM070560. We thank Dr. Qingfen Li for proof reading.
Addendum to:
References
- Blobel G, Dobberstein B. Transfer to proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol 1975; 67:852 - 862
- St. Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol 2005; 6:363 - 375
- Liao G, Ma X, Liu G. An RNA-zipcode-independent mechanism that localizes Dia1 mRNA to the perinuclear ER through interactions between Dia1 nascent peptide and Rho-GTP. J Cell Sci 2011; 124:589 - 599
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell 2005; 18:273 - 281
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 2005; 435:513 - 518
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol 2007; 178:193 - 200
- Brown MD, Sacks DB. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol 2006; 16:242 - 249
- Brandt DT, Grosse R. Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep 2007; 8:1019 - 1023
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol 1981; 91:551 - 556
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 2011; 331:586 - 589
- Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol Cell 2007; 99:475 - 487
- Zhang G, Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with cotranslational folding. Curr Opin Struct Biol 2011; 21:25 - 31
- Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci 2005; 118:2425 - 2433
- Liao G, Simone B, Liu G. Mis-localization of Arp2 mRNA impairs persistence of directional cell migration. Exp Cell Res 2011; 317:812 - 822
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol 2004; 6:820 - 830
- Bola B, Allan V. How and why does the endoplasmic reticulum move?. Biochem Soc Trans 2009; 37:961 - 965