Abstract
Within prokaryotes cyanobacteria represent one of the oldest and morphologically most diverse phyla on Earth. The rise of oxygen levels in the atmosphere 2.32-2.45 billion years ago is assigned to the photosynthetic activity of ancestors from this phylum. Subsequently cyanobacteria were able to adapt to various habitats evolving a comprehensive set of different morphotypes. In a recent study we showed that this evolution is not a gradual transition from simple unicellular to more complex multicellular forms as often assumed. Instead complexity was lost several times and regained at least once. An understanding of the genetic basis of these transitions would be further strengthened by phylogenomic approaches. However, considering that new methods for phylogenomic analyses are emerging, it is unfortunate that genomes available today are comprised of an unbalanced sampling of taxa. We propose avenues to remedy this by identifying taxa that would improve the representation of phylogenetic diversity in this phylum.
Cyanobacteria are a group of prokaryotes that obtain their energy through oxygenic photosynthesis. The rise of oxygen levels in Earth's atmosphere 2.32–2.45 billion years ago, often referred to as the “Great Oxygenation Event,”Citation1 provides the first indirect evidence for the presence of cyanobacteria. Based on their morphology, cyanobacteria represent one of the most diverse prokaryotic phyla. They have been divided into five different morphological sections.Citation2 Sections I and II comprise unicellular species, whereas sections III, IV and V consist of multicellular species. Sections IV and V evolved terminal differentiation and therefore represent the most morphologically complex cyanobacteria. Fossil records of cyanobacteria are well established and can be found in 2.0 billion year old rock formations.Citation3 In a recently published study, we traced the origin of multicellularity in cyanobacteria, and found a pattern of frequent morphological transitions.Citation4 A fast heuristic maximum likelihood approach made it possible to reconstruct a phylogeny of cyanobacteria for over a thousand taxa based on 16S rRNA sequences. A smaller subset of cyanobacteria representing full morphological and genetic diversity was used for the maximum likelihood and Bayesian reconstruction of ancestral character states. These analyses demonstrated that all cyanobacteria share a unicellular most recent common ancestor. However, multicellularity evolved very early in the cyanobacterial lineage. Evolution and divergence from the ancient multicellular lineage led to the richness of morphotypes manifested by cyanobacteria today. In the course of the cyanobacterial diversification process, complexity in the form of multicellular growth was lost at least five times. It was also regained at least once. Furthermore, by comparing the results to the prokaryotic fossil record, it was possible to elucidate that multicellularity evolved in cyanobacteria earlier than 2.0 billion years ago, possibly prior to the “Great Oxygenation Event.” Several multicellular prokaryotic fossils that are suggestive of cyanobacterial morphotypes have been found in rock formations from the Archean Eon, which corroborates our conclusions.Citation5
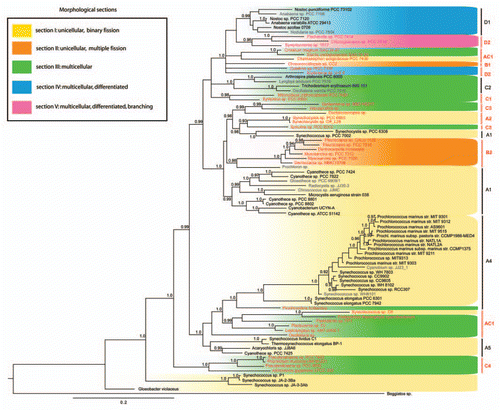
For this addendum, we reconstructed two cyanobacterial phylogenies inferred with the general substitution model (GTR + G + I) from 16S rRNA sequences, and emphasize the position of fully sequenced genomes in the sample. displays a phylogenetic tree reconstructed from 1,261 cyanobacterial taxa (1,206 sites) using maximum likelihood as implemented in RAxML v7.0.4.Citation6 Branch supports represent bootstrap values calculated from 100 re-samplings of the dataset. shows the phylogenetic tree built with a Bayesian approach as implemented in MrBayes v3.1.Citation7 Two Metropolis-coupled MCMC runs were performed over 20,000,000 generations, sampling trees and parameters every 1,000th generation. Convergence of the parameters was checked using AWTYCitation8 and the first 50% of trees (namely 10,000) were discarded as burn-in. The trees were partitioned in groups (A1–A5; AC1; B1, B2; C1–C4; D1, D2) as previously described in reference Citation4. Upon examining the figures, an over-representation of genome data from unicellular cyanobacterial taxa is striking. Out of 41 fully sequenced cyanobacterial strains, 35 are single celled. All fully sequenced unicellular taxa belong to morphological section I, and are located in groups A1–A5 in our phylogeny. The six multicellular taxa for which full genome data are available belong to morphological sections III and IV, which are located in groups D1 and C2 ( and ). Although it seems that the fully sequenced strains belong to taxa-rich phylogenetic groups, this pattern could result from a bias in sampling or cultivation success. Therefore the apparent size of groups containing cultivated cyanobacteria (in terms of the number of taxa) does not necessarily represent its actual size in nature. In the reconstructed trees the taxonomic sampling from smaller groups is likely to contribute demonstrably to increased phylogenetic diversity (PD). Unfortunately no sequence data are available for those groups (AC1, B1, B2, C1, C3, C4 and D2). Morphological sections II and V lack genome data completely. shows a well resolved phylogenetic reconstruction of cyanobacterial taxonomy, highlighting in red the species recommended for future genome projects. Multicellular species are found in groups D1, D2, C1, C2, C3, C4 and AC1, but only six species from two of these groups (D1, C2) are completely sequenced. Special attention should be brought to multicellular species from groups AC1 and C4 which branch closer to the outgroup () and therefore would be valuable for exploring the origin of multicellularity. Furthermore, genomes for future sequencing can be rigorously selected in order to maximize both the phylogenetic and morphological diversity. This can for example be done using a simple “greedy” algorithm,Citation9 by iteratively choosing the best immediate taxon (i.e., those adding most PD) based on the phylogeny depicted here.
State of the art methods for supermatrix and supertree construction aim to infer the species phylogeny from thousands of taxa and multiple genomic loci.Citation10 Although for prokaryotes inferring a single species phylogeny may be particularly hampered by frequent lateral gene transfer, recombination and species delimitation problems.Citation11 Nonetheless the availability of better genomics data will make it possible to study the distribution of individual gene histories, from which the evolutionary mechanisms and their patterns may be inferred. Developments in statistical and computational methodology offer improved alignment strategiesCitation12 and enable inference in the presence of recombination, incomplete lineage sorting and lateral gene transfer. In addition, the natural lineage heterogeneity may now be studied at the genomic scale using non-stationary or nonreversible Markovian models,Citation13 leading to the better understanding of the biological forces driving lineage specific biases such as those affecting the evolutionary rates and molecular composition in diverse groups.
A more representative sampling of cyanobacterial species would provide a basis for future phylogenomic analyses that would offer insights into the early history of cyanobacteria. This would not only strengthen and further test the results of the study discussed here, but also enable others to identify and reconstruct the evolution of a wide set of genes that have been responsible for morphological and biochemical adaptations in this phylum.
Figures and Tables
Figure 1 Phylogenetic tree of 1,261 cyanobacterial taxa reconstructed by maximum likelihood from 16S rRNA sequences. Morphological sections are marked by color. Fully sequenced strains are indicated on the tree by their taxonomic names. Phylogenetic groups (A1–A5; B1, B2; C1–C4; D1, D2) have been described elsewhere. Four groups for which no genome data are available at present are marked red. Taxa have been selected as previously described in reference Citation4, with the addition of 41 taxa for which full genome sequences are available.
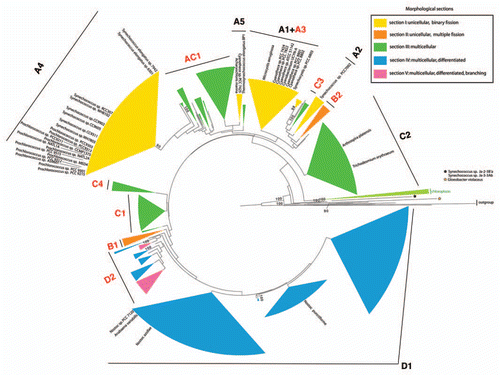
Figure 2 Phylogenetic tree of a cyanbacterial subset representing the full morphological and genomic diversity. The tree was reconstructed from 16S rRNA sequences using a Bayesian approach. Colors describe different morphological sections. Fully sequenced taxa are shown in black. Shown in red are taxa of groups where no genome sequence data are available at present, and which we recommended for genome sequencing. Taxa have been selected as previously described in reference Citation4, with the addition of 41 taxa for which full genome sequences are available.
Addendum to:
References
- Bekker A, Holland HD, Wang PL, Rumble D, Stein HJ, Hannah JL, et al. Dating the rise of atmospheric oxygen. Nature 2004; 427:117 - 120; PMID: 14712267; http://dx.doi.org/10.1038/nature02260
- Castenholz RW, Phylum BX. Garrity G, Boone DR, Castenholz RW. Cyanobacteria. Oxygenic Photosynthetic Bacteria. Bergey's Manual of Systematic Bacteriology. Volume 1: The Archaea and the Deeply Branching and Phototropic Bacteria 2001; New York, US Springer-Verlag 473 - 600
- Amard B, Bertrand-Sarfati J. Microfossils in 2000 Ma old cherty stromatolites of the Franceville Group, Gabon. Precambrian Res 1997; 81:197 - 221; http://dx.doi.org/10.1016/S0301-9268(96)00035-6
- Schirrmeister BE, Antonelli A, Bagheri HC. The origin of multicellularity in cyanobacteria. BMC Evol Biol 2011; 11:45; PMID: 21320320; http://dx.doi.org/10.1186/1471-2148-11-45
- Wacey D. Early Life on Earth (A practical guide) 2009; Springer-VScience+Buisness Media BV
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22:2688 - 2690; PMID: 16928733; http://dx.doi.org/10.1093/bioinformatics/btl446
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19:1572 - 1574; PMID: 12912839; http://dx.doi.org/10.1093/bioinformatics/btg180
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. Bioinformatics 2008; 24:581 - 583; PMID: 17766271; http://dx.doi.org/10.1093/bioinformatics/btm388
- Pardi F, Goldman N. Species Choice for Comparative Genomics: Being Greedy Works. PLoS Genet 2005; 1:71; PMID: 16327885; http://dx.doi.org/10.1371/journal.pgen.0010071
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nature Rev Genet 2005; 6:361 - 375; PMID: 15861208; http://dx.doi.org/10.1038/nrg1603
- Doolittle WF. Uprooting the tree of life. Scientific American 2000; 282:90 - 95
- Anisimova M, Cannarozzi GM, Liberles DA. Finding the balance between the mathematical and biological optima in multiple sequence alignment. Trends Evol Biol 2010; 2:7; http://dx.doi.org/10.4081/eb.2010.e7
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M. Parallel adaptations to high temperatures in the Archaean eon. Nature 2008; 456:942 - 945; PMID: 19037246; http://dx.doi.org/10.1038/nature07393