Abstract
Phospholipases C (PLCs) that hydrolyze inositol phospholipids regulate vital cellular functions in both eukaryotic and prokaryotic organisms. The PLC superfamily consists of eukaryotic phosphoinositide-specific PLCs (PI-PLCs), bacterial PLCs and trypanosomal PLCs.1 PI-PLCs hydrolyze phosphatidylinositol 4,5-bisphosphate (PtdIns4,5P2) to produce inositol 1,4,5-trisphosphate (Ins1,4,5P3) and constitute a hallmark feature of eukaryotic cells. In metazoa, this reaction is coupled to receptor signaling via specific PI-PLC isoforms and results in acute increase of cytosolic Ca2+ levels by Ins1,4,5P3-sensitive Ca2+ channels (IP3-receptors, IP3Rs).2 A striking result of many studies so far has been the presence of a single PI-PLC gene in all unicellular eukaryotes investigated, as opposed to expansion of PI-PLC isoforms in metazoa;3 this has suggested that a single housekeeping PI-PLC represents an archetypal and simplified form of PI-PLC signaling.3 Several studies however have noted a unique expansion of PI-PLC/IP3R pathway components in ciliates.4,5 In a recent paper we showed the presence of multiple functional PI-PLC genes in Tetrahymena thermophila and biochemical characterization, pharmacological studies and study of their expression patterns suggested that they are likely to serve distinct non-redundant roles.4 In this report we discuss these studies and how they advance our understanding of PI-PLC functions in ciliates.
Unique Combinations of PLC Genes in Ciliates
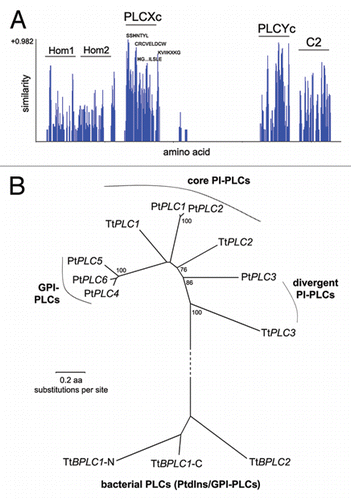
Biochemical probing of PLC activities in homogenate and subcellular fractions from two Tetrahymena species revealed the presence of two distinct PLC activities utilizing PtdIns and PtdIns4,5P2. PtdIns was utilized by a Ca2+-insensitive/inhibited PLC, while PtdIns4,5P2 was hydrolyzed by a membrane-associated, low micromolar Ca2+-activated PLC. The latter activity was inhibited in vitro by the PI-PLC inhibitor U73122, indicating a bona-fide eukaryotic PI-PLC. These two activities were attributed to two sets of genes identified in the Tetrahymena genome that correspond to bacterial PtdIns-PLCs (TtBPLC1–2) and eukaryotic PI-PLCs (TtPLC1–3).Citation4 In an another ciliate, Paramecium tetraurelia, six eukaryotic PI-PLC genes are present revealing a unique expansion in ciliates (for the sake of simplicity we have utilized the assignment in ref. Citation4 instead of that in ref. Citation6 for Paramecium PLCs). Phylogenetic analysis indicates further diversity within the ciliate clade. The core of ciliate PI-PLCs appears to be composed of TtPLC1, TtPLC2, PtPLC1 and PtPLC2 ( and B). Two genes, TtPLC3 and PtPLC3, are evolutionarily divergent and, based on analysis of their PLCXc domains, they cannot be consistently assigned to a specific PI-PLC group (see Fig. 5C and Sup. Fig. S2D in ref. Citation4 for details). TtPLC3 is a novel inactive PI-PLC isoform resembling PLC-L/PRIP proteins identified only in metazoa,Citation4 but PtPLC3 apparently codes for an active enzyme.Citation6 Adding to the diversity within ciliate PI-PLCs, three Paramecium genes (PtPLC4–6) form a distinct groupCitation4,Citation6 () and they have been shown to be specifically involved in GPI anchor cleavage (a typical characteristic of bacterial and trypanosomal PLCs) rather than PtdIns4,5P2 hydrolysis.Citation6 In Tetrahymena, a bacterial PLC gene, TtBPLC1, codes for a catalytically competent PtdIns-PLC, and may correspond to a secreted activity that participates in extracellular digestion of PtdIns;Citation4 given the well-established activity of other bacterial PLCs in hydrolyzing GPI-anchored proteins,Citation1 it may be a possible candidate for cleavage of GPI-anchors in Tetrahymena. In conclusion, although there is significant diversity of PLC genes within these two ciliates (), three PI-PLC genes in Tetrahymena (TtPLC3/PRIP apparently in an indirect manner),Citation4 and at least two in Paramecium (since PtPLC2 is postulated to be a non-functional gene),Citation6 are likely to participate exclusively in PtdIns4,5P2 hydrolysis and hence generation of Ins1,4,5P3 in vivo ().
Occurrence of PI-PLC-Generated Ins1,4,5P3 in Ciliates
Tetrahymena PI-PLC activity is actively involved in hydrolysis of PtdIns4,5P2 in vivo, since short-term treatment with U73122 results in significant upregulation of PtdIns4,5P2 levels.Citation4 Furthermore, GTPγS exerted a modest activation of PI-PLC activity in vitro indicating that GTPases are likely to play a regulatory (activating) role.Citation4 These data advance significantly our knowledge and validate our pharmacological tools for dissecting the roles of ciliate PI-PLCs in vivo. Nevertheless, we still lack definite evidence for stimulus-induced formation of Ins1,4,5P3 per se in either Tetrahymena or Paramecium. Use of Dowex chromatography for detection of Ins1,4,5P3 in Dictyostelium resulted in erroneous assumptions due to the presence of other co-eluted inositol trisphosphates,Citation7 and it has yielded inconsistent results in detection of inositol-labeled Tetrahymena InsP3 compounds in vivo (our data). Furthermore, an early study utilizing HPLC in Paramecium has failed to present evidence for the presence of Ins1,4,5P3.Citation8 Nevertheless, the use of a standard Ins1,4,5P3 mass assay has yielded positive results in Tetrahymena, where the abundance of Ins1,4,5P3 was estimated to be approximately 10 pmol/106 cells (Leondaritis G and Galanopoulou D, unpublished data). This value is comparable to the abundance of PtdIns4,5P2 which is deduced to be in the range of 100 pmol/106 cells (estimation based on the mass quantification of PtdIns and [3H] myo-inositol labeling, assuming isotopic equilibrium between all phosphoinositide pools).Citation9 These compelling results however are faced with an early Paramecium study suggesting that the Ins1,4,5P3 mass assay may also detect other inositol trisphosphate isomers produced by a InsP6-phosphatase activity (in vitro), with equal sensitivity.Citation8 Apparently, resolving this issue will be important in understanding the extent of PI-PLC-generated Ins1,4,5P3 in vivo in either Tetrahymena or Paramecium.
Roles of PI-PLCs in Ciliates
In Tetrahymena, comparative studies of expression patterns and activity in three strains suggested a high level of co-ordination between expression levels of TtPLC2 (and probably TtPLC1) and total PI-PLC activity.Citation4 What would be then the role of PI-PLC-generated Ins1,4,5P3 in Tetrahymena? Recent studies on the presence and functionality of IP3R genes in Paramecium,Citation5,Citation10 also present in Tetrahymena as well,Citation5 suggest that Ins1,4,5P3 may indeed regulate Ca2+ homeostasis in ciliates. Study of a specific Paramecium IP3R (CRC-II-1a,b/IP3RN1,2) has shown that it regulates Ca2+ mobilization in a restricted subcellular location, the contractile vacuole complex,Citation10 and it may have a latent role in regulation of basal Ca2+ homeostasis.Citation5 The fact that uncaging of Ins1,4,5P3 induces only short and strictly localized Ca2+ mobilization in Paramecium suggests that it is unlikely to promote global and large mobilization of intracellular Ca2+.Citation10,Citation11 Instead, a compartmentalized function of PI-PLC-generated Ins1,4,5P3 as a Ca2+-mobilizing agent in a localized fashion in the contractile vacuole complex or other—yet unidentified—subcellular locations, rich in one or more of the approximately 16 IP3R-related CRC channels in ParameciumCitation5 (and the like in Tetrahymena), seems more plausible with the current knowledge. In addition, the well-correlated expression patterns of TtPLC3/PRIP and TtPLC2 (but not TtPLC1) during Tetrahymena conjugation,Citation4 point to a specific role for PI-PLC-generated Ins1,4,5P3 in this process during which Ca2+ dynamics and requirements may be quite different. Furthermore, circumstantial evidence from T. thermophila mutant strainsCitation4 and the phenotype of CRC-II-1a,b silencing in ParameciumCitation5 point to the possibility that PI-PLC activity may impinge on secretory pathways, an aspect that would be interesting to study in detail in the future.
A last point deserves further attention. In S. cerevisiae, PI-PLC-generated Ins1,4,5P3 is the substrate for the synthesis of higher inositol phosphates that are recognized as important players in transcription and chromatin remodeling.Citation2,Citation12 The first step is catalyzed by a ubiquitous eukaryotic multifunctional enzyme known as IPK2/IPMK, to yield Ins(1,4,5,6)P4 and subsequently Ins(1,3,4,5,6)P5.Citation12 In Tetrahymena, an intact IPK2/IPMK-like catalytic domain is encoded by the Thd2 gene, a class II histone deacetylase.Citation13 Cells defective in Thd2 were shown to exhibit chromatin and cytological phenotypes indicative of a role for Thd2 in chromatin maturation including the proteolytic processing of histone H3 in micronucleous,Citation13 but the involvement of the IPK2/IPMK domain is unknown. Since TtPLC1–3 have nuclear export signals and TtPLC3 possesses additional nuclear localization signals,Citation4 it would be interesting to examine the possibility of co-localization with Thd2 in Tetrahymena nuclei.Citation13
In conclusion, several recent studies have highlighted the presence of PI-PLCs in the ciliates Tetrahymena and Paramecium and future experiments along these lines may well unravel novel functions and novel modes of regulation of PI-PLC signaling in eukaryotes. The connection with Ca2+ homeostasis seems quite solid, although several parts of the puzzle are still uncertain (some are also discussed in ref. Citation10 and Citation11). The characterization of the phenotypes of PLC-deficient cells will add significantly to our understanding of evolution of PI-PLC signaling in eukaryotic organisms and will probably require expertise in diverse fields of ciliate biology.
Abbreviations
| CRC | = | calcium release channel |
| HPLC | = | high-performance liquid chromatography |
| Ins1,4,5P3 | = | inositol-1,4,5-trisphosphate |
| IP3R | = | inositol-1,4,5-trisphosphate receptor |
| IPK2/IPMK | = | inositol phosphate kinase 2/inositol phosphate multikinase |
| PI | = | phosphoinositide |
| PLC | = | phospholipase C |
| PtdIns | = | phosphatidylinositol |
| PtdIns4,5P2 | = | phosphatidylinositol-4,5-bisphosphate |
Figures and Tables
Figure 1 Ciliate PI-PLC genes. (A) Similarity graph of the consensus obtained after multiple sequence alignment of full length TtPLC1–3 and PtPLC1–6. Regions of significant similarity correspond primarily to PLCXc, PLCYc and C2 domains. Key features in the PLCXc domain are indicated as well (see Sup. Fig. 2C in ref. Citation4 for a detailed description). The gap between PLCXc and PLCYc domains is due to the large TtPLC3/PRIP X–Y linker. Hom 1 (homology 1) region corresponds to the predicted PH domain of TtPLC3/PRIP (reviewed in ref. Citation4). Interestingly, the respective region in PtPLC2 (31–136 aa) is retrieved as a putative PH domain in SMART database and blastp searches against all Tetrahymena and Paramecium genes using this PtPLC2 region recovers PtPLC1 (e-value = e−57), TtPLC1–2 (e-value= e−6 − e−9) and TtPLC3/PRIP (e-value = e−3) as the best scoring hits. The Hom 2 (homology 2) region roughly corresponds to the predicted EF hand motifs in TtPLC1–3 and PtPLC1–2 (reviewed in ref. Citation4 and Citation6, respectively). (B) The indicated evolutionary relationships were inferred from alignments of the PLCXc domain using the Neighbor-Joining method while the evolutionary distances were computed using the Poisson correction method (MEGA 4.0 software, as in ref. Citation4). PtPLC1 and 2 and PtPLC4 and 6 are pairs of paralogous genes from the last whole genome duplication in P. tetraurelia (reviewed in ref. Citation6), while TtPLC3/PRIP is the most divergent ciliate PLC. Tetrahymena BPLC1 and BPLC2 PLCXc domains were positioned as an outgroup to the rest PLCs. Bootstrap values from 1,000 replicates (higher than 75%) are indicated near corresponding branches. Gene locus tags for ciliate PLCs are: TtPLC1, TTHERM_00486470; TtPLC2, TTHERM_00238850; TtPLC3/PRIP, TTHERM_00085110; PtPLC1, (PLC3 in ref. Citation6), GSPATT00031253001; PtPLC2 (PLC5 in ref. Citation6), GSPATT00029973001; PtPLC3 (PLC1 in ref. Citation6), GSPATT00008002001; PtPLC4, GSPATT00031342001; PtPLC5 (PLC2 in ref. Citation6) GSPATT00034681001; PtPLC6, GSPATT00030070001; TtBPLC1, TTHERM_00426140; TtBPLC2, TTHERM_00348190.
Acknowledgments
We thank our students, T. Sarri, A. Efstathiou and Dr. I. Dafnis for their help during distinct phases of this project. We also thank Professor W.F. Boss and Dr. I.Y. Perera, Department of Plant Biology, NCSU, USA, for their help with Ins1,4,5P3 assay. This work was partially supported by a University of Athens grant (KA 70/4/8779).
Addendum to:
References
- Heinz DW, Essen LO, Williams RL. Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J Mol Biol 1998; 275:635 - 650
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol 2001; 2:327 - 338
- Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol 2008; 9:151 - 161
- Leondaritis G, Sarri T, Dafnis I, Efstathiou A, Galanopoulou D. Biochemical and genetic evidence for the presence of multiple phosphatidylinositol- and phosphatidylinositol-4,5-bisphosphate-specific phospholipases C in Tetrahymena. Eukaryot Cell 2011; 10:412 - 422
- Ladenburger EM, Sehring IM, Korn I, Plattner H. Novel types of Ca2+-release channels participate in the secretory cycle of Paramecium cells. Mol Cell Biol 2009; 29:3605 - 3622
- Kloppel C, Muller A, Marker S, Simon M. Two isoforms of eukaryotic phospholipase C in Paramecium affecting transport and release of GPI-anchored proteins in vivo. Eur J Cell Biol 2009; 88:577 - 592
- van Haastert PJM, de Vries MJ, Penning LC, Roovers E, van der Kaay J, Erneux C, van Lookeren Campagne MM. Chemoattractant and guanosine 5′-[γ-thio]triphosphate induce the accumulation of inositol-1,4,5-trisphosphate in Dictyostelium cells that are labelled with [3H]inositol by electroporation. Biochem J 1989; 258:577 - 586
- Freund WD, Mayr GW, Tietz C, Schultz JE. Metabolism of inositol phosphates in the protozoan Paramecium: Characterization of a novel inositol-hexakisphosphate-dephosphorylating enzyme. Eur J Biochem 1992; 207:359 - 367
- Leondaritis G, Galanopoulou D. Characterization of inositol phospholipids and identification of a mastoparan-induced polyphosphoinositide response in Tetrahymena pyriformis. Lipids 2000; 35:525 - 532
- Ladenburger EM, Korn I, Kasielke N, Wassmer T, Plattner H. An Ins(1,4,5)P3 receptor in Paramecium is associated with the osmoregulatory system. J Cell Sci 2006; 119:3705 - 3717
- Plattner H, Sehring IM, Schilde C, Ladenburger EM. Pharmacology of ciliated protozoa-drug (in)sensitivity and experimental drug (ab)use. Int Rev Cell Mol Biol 2009; 273:163 - 218
- Shears SB. How versatile are inositol phosphate kinases?. Biochem J 2004; 377:265 - 280
- Smith JJ, Torigoe SE, Maxson J, Fish LC, Wiley EA. A class II histone deacetylase acts on newly synthesized histones in Tetrahymena. Eukaryot Cell 2008; 7:471 - 482