Abstract
In the oral cavity, gingival epithelial cell (GEC) layers function as an innate host defense system to prevent intrusion by periodontal bacteria. Nevertheless, Porphyromonas gingivalis, the most well-known periodontal pathogen, can enter GECs and pass through the epithelial barrier into deeper tissues. An intracellular location is considered advantageous for bacteria to escape from immune surveillance by the host as well as antibiotic pressure, leading to intracellular persistence, multiplication, and dissemination to adjacent tissues. P. gingivalis are invaginated by gingival epithelial cells via the endocytic pathway, and some intracellular bacteria are sorted to lytic compartments, including autolysosomes and late endosomes/lysosomes, while a considerable number of the remaining organisms are sorted to Rab11- and RalA-positive recycling endosomes, followed by bacterial exit from the cells. Exited bacteria can re-enter fresh cells. However, dominant negative forms and RNAi-knockdown of Rab11, RalA, and exocyst complex subunits (Sec5, Sec6, and Exo84) significantly disturb the exit of P. gingivalis. These are the first known results to show that the endocytic recycling pathway mediates bacterial exit from infected cells to neighboring cells and may provide important information regarding the exit mechanisms of various invasive pathogens.
Marginal periodontitis, one of the most common infectious diseases seen in humans, is characterized by gingival inflammation, as well as loss of connective tissue and bone from around the roots of the teeth, which lead to eventual tooth exfoliation.Citation1 A variety of dental plaque-forming bacteria were shown to penetrate the epithelium of periodontal pockets in individuals with advanced periodontitis, while periodontal bacteria were found to enter periodontal cells. Casadevall noted that the intracellular infection process for most invasive microorganisms can be divided into 4 phases; adhesion, entry, survival and exit.Citation2 In addition, it was shown that exit from infected cells enables further penetration into host tissues and provides invading bacteria a means to control their populations in infected tissues, while also allowing for persistent infection. Those results indicated that initial (direct or indirect) interactions between bacterial adhesins/invasins and cellular receptors trigger a cascade of signals, including protein phosphorylation and activation of cytoskeleton components to engulf the bacterium with a phagocytic cup, followed by bacterial internalization. Porphyromonas gingivalis also exploits cellular endocytosis to enter cells, including epithelial and endothelial cells (), which is mediated by the interaction between bacterial fimbriae and cellular α5β1 integrin.Citation3
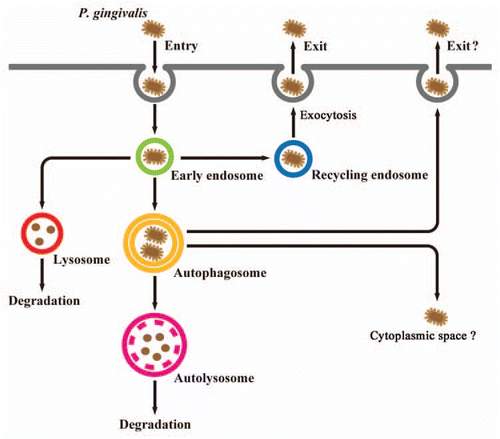
Other studies have shown that intracellular P. gingivalis is able to exit from primarily infected host cells into intercellular space and enter new host cells, which enables further penetration into host tissues in a trans-cellular manner.Citation4,Citation5 Thus, P. gingivalis is suggested to use such a mechanism to control its population in infected tissues, yet allow for persistent infection. We have reported that the cellular recycling pathway is exploited by intracellular P. gingivalis to allow the bacteria to exit from infected cells to neighboring cells in a mechanism of cell-to-cell spreading ().Citation6 P. gingivalis was internalized with early endosomes positive for the FYVE domain of EEA1 (an early endosome marker) and transferrin receptor, and about half of the intracellular bacteria were found to be sorted to lytic compartments, including autolysosomes and late endosomes/lysosomes, while a considerable number of the remaining organisms were sorted to Rab11- and RalA-positive recycling endosomes, followed by the bacterial exit. Inhibition experiments revealed that bacterial exit is dependent on actin polymerization, lipid rafts and microtubule assembly, while dominant negative forms and RNAi-knockdown of Rab11, RalA and exocyst complex subunits (Sec5, Sec6 and Exo84) significantly disturbed the exit of P. gingivalis. It should be noted that intracellular bacteria were found to be sorted to three different destinations within the same cells. The intracellular trafficking dynamics of this bacterium is composed of multiple pathways and our results imply the existence of unknown mechanism(s) that alternate among multiple sorting pathways destined for autophagy, lysosomes or the recycling pathway.
It has not been clearly elucidated how P. gingivalis traffics within infected cells. Intracellular P. gingivalis reportedly localizes in various cellular compartments, such as the cytoplasm,Citation7–Citation10 endosomes,Citation8–Citation12 and autophagosomes,Citation11–Citation13 which are distinctive double-membraned vacuoles that are part of the autophagic pathway, with autophagosome localization noted to occur in endothelial and smooth muscle cells,Citation11 but not in gingival epithelial cells.Citation7 On the other hand, no bacteria were found in the cytoplasmic spaces of endothelial cells.Citation11,Citation12 We showed that P. gingivalis is internalized in early endosomes and about half of the intracellular bacteria were found to be sorted to lytic compartments, including autolysosomes and late endosomes/lysosomes.Citation6
Intracellular trafficking of P. gingivalis may depend on the phenotypic and genotypic heterogeneity of the host cells. In endothelial cells, P. gingivalis was reported to traffic ultimately to autophagosomes.Citation10,Citation13 Once in these membrane compartments, the bacteria block fusion with lysosomes and likely use protein debris trafficked through the autophagic pathway as their nutrient source. P. gingivalis may persist in these autophagosome vacuoles, and inflammation and cell damage can result from accumulations of high loads of intracellular organisms. In our study, wortmannin (autophagy inhibitor) was effective to reduce the number of intracellular bacteria,Citation6 indicating the possibility that autophagy allows intracellular P. gingivalis to survive. However, the role of autophagy in survival of intracellular P. gingivalis has not been well defined and is currently being examined in our laboratory.
P. gingivalis is able to enter and exit periodontal cells using a sophisticated strategy that enables the expansion of infection in periodontal tissues. Analyses of bacterial strategies related to the inter-cellular persistence, dissemination and fate of P. gingivalis are continuing, though only a portion of the picture is currently known. Additional molecular dissection studies are required to better understand the intracellular lifestyle of periodontal bacteria.
Figures and Tables
Figure 1 Entry of P. gingivalis into human gingival epithelial cells. Human gingival epithelial cells were incubated with P. gingivalis strain ATCC 33277 for 15 min and bacterial entry was observed using a scanning electron microscope. P. gingivalis is captured by cellular pseudopodia and internalized via an endocytic pathway.

Figure 2 Proposed model of P. gingivalis trafficking in human gingival epithelial cells. P. gingivalis organisms are initially localized within endocytic vacuoles (early endosomes) after entry. Some bacteria are routed to late endosomes, then subsequently sorted to lysosomes for degradation. Other bacteria promote their own entry into the autophagic pathway by bacterial escape from endosomes or fusion of endosomes with autophagosomes, and are then sorted to autolysosomes, which are formed by fusion of autophagosomes with lysosomes for degradation. Some intracellular P. gingivalis organisms are able to escape from endosomes to the recycling pathway. Subsequently, the bacteria exit from primarily infected host cells, which may enable further penetration of host tissues in a trans-cellular manner. It remains unknown whether P. gingivalis can escape the autophagic machinery to cytoplasmic space or extracellular milieu.
Addendum to:
References
- Lamont RJ, Yilmaz O. In or out: the invasiveness of oral bacteria. Periodontol 2002; 30:61 - 59
- Casadevall A. Evolution of intracellular pathogens. Annu Rev Microbiol 2008; 62:19 - 33
- Amano A. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front Biosci 2007; 12:3965 - 3974
- Li L, Michel R, Cohen J, Decarlo A, Kozarov E. Intracellular survival and vascular cell-to-cell transmission of Porphyromonas gingivalis. BMC Microbiol 2008; 8:26
- Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun 2006; 74:703 - 710
- Takeuchi H, Furuta N, Morisaki I, Amano A. Exit of intracellular Porphyromonas gingivalis from gingival epithelial cells is mediated by endocytic recycling pathway. Cell Microbiol 2011; 13:677 - 691
- Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun 1995; 63:3878 - 3885
- Sandros J, Papapanou P, Dahlén G. Porphyromonas gingivalis invades oral epithelial cells in vitro. J Periodontal Res 1993; 28:219 - 226
- Papapanou PN, Sandros J, Lindberg K, Duncan MJ, Niederman R, Nannmark U. Porphyromonas gingivalis may multiply and advance within stratified human junctional epithelium in vitro. J Periodontal Res 1994; 29:374 - 375
- Duncan MJ, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun 1993; 61:2260 - 2265
- Dorn BR, Dunn WA Jr, Progulske-Fox A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect Immun 2001; 69:5698 - 5708
- Yamatake K, Maeda M, Kadowaki T, Takii R, Tsukuba T, Ueno T, et al. Role for gingipains in Porphyromonas gingivalis traffic to phagolysosomes and survival in human aortic endothelial cells. Infect Immun 2007; 75:2090 - 100
- Bélanger M, Rodrigues PH, Dunn WA Jr, Progulske-Fox A. Autophagy: a highway for Porphyromonas gingivalis in endothelial cells. Autophagy 2006; 2:165 - 170