Abstract
Centrosomes are microtubule-organizing centers that nucleate spindle microtubules during cell division. In budding yeast, the centrosome, often referred to as the spindle pole body, shares structural components with the centriole, the central core of the animal centrosome. The parental centrosome is duplicated when DNA replication takes place. Like sister chromatids tethered together by cohesin, duplicated centrosomes are linked and then separate to form the bipolar spindle necessary for chromosome segregation. Recent studies have shown that cohesin is also localized to the animal centrosome and is perhaps directly involved in engaging paired centrioles. Here we discuss the potential role of cohesin in mediating spindle-pole-body cohesion in the context of yeast meiosis. We propose that the coordination of chromosome segregation with centrosome cohesion and duplication is mediated by the antagonistic interaction between the Aurora kinase and the Polo kinase and that the role of cohesin in centrosome regulation appears to be indirect in budding yeast.
The microtubule-organizing center in yeast is often referred to as the spindle pole body (SPB) and shares structural and functional components with the animal centrosome.Citation1 We focus here on the dynamics of the meiotic yeast SPB, which provide an unparalleled system for the dissection of centrosome structure and function. Meiosis produces gametes that contain only half of the parental genome. During meiosis, homologs and sister chromatids separate sequentially in two continuous cell divisions after one round of DNA replication. This unique pattern of chromosome segregation depends on the formation of a bipolar spindle that separates homologs in meiosis I and on the simultaneous formation of two independent spindles that permit sister-chromatid separation in meiosis II (). The SPB/centrosome nucleates spindle microtubules. During DNA replication at interphase I, the parental centrosome is duplicated to form two sister centrosomes. Sister centrosomes are duplicated again in the absence of DNA replication at the end of meiosis I, called interphase II, and establish two spindles in meiosis II ().
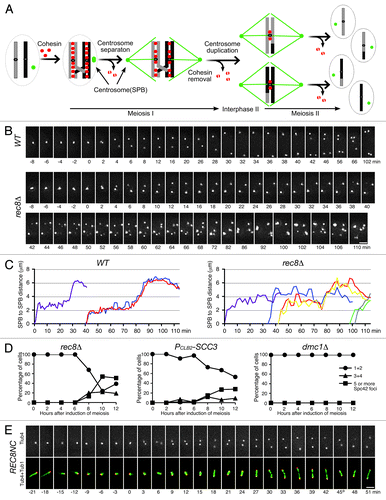
Figure 1. Requirement for cohesin for SPB cohesion and duplication during yeast meiosis. (A) A schematic diagram showing spindle pole body (SPB, centrosome) and chromosome segregation during meiosis. Chromosomes are shown as gray and black bars; SPB, green dots; cohesin, red dots; microtubules, green lines. (B) Live-cell fluorescence microscopy showing SPB dynamics in wild type (WT) and rec8Δ cells during yeast meiosis. SPBs are marked by Spc42‑GFP. Projected images from eight z-stacks with 1‑μm optical sectioning are shown. Exposure time for each optical section was 100 ms. Time zero is defined as the point of SPB separation in meiosis I. Time lapse was 2 min. Note that SPBs are only loosely connected before separation in rec8Δ cells. (C) Pole-to-pole distance from WT and rec8Δ cells as shown in B. MI spindle, purple; MII spindle, other colors. (D) Live-cell fluorescence microscopy showing SPB and microtubule spindle dynamics during yeast meiosis in a REC8NC (Rec8-noncleavableCitation19) cell. SPBs are marked by Tub4 (g‑tubulin)-GFP, microtubules by Tub1 (a‑tubulin)-mApple. Projected images from seven z-stacks with 1-μm optical sectioning are shown. Exposure time for each optical section was 60 ms. Time zero was defined as in B. Time-lapse was 3 min. Time in minutes is shown below each frame. * indicates the time of spindle recovery after breakage. Bars, 2 μm.
Cohesin, a multisubunit protein complex, is required for generating sister-chromatid cohesion after DNA replication. Originally identified in budding yeast, cohesin is composed of four subunits called Smc1, Smc3, Mcd1/Scc1/Rad21, and Scc3/SA/STAG.Citation2,Citation3 Rec8 largely replaces Mcd1 and is the only meiosis-specific cohesin subunit in budding yeast. Intriguingly, cohesin also localizes to the animal centrosome,Citation4-Citation6 and siRNA-mediated depletion of cohesin subunit Rad21 causes premature separation of paired centrioles,Citation7 the core of the centrosome. Separase, which cleaves cohesin, is necessary for centriole disengagement and licenses centrosome duplication.Citation8,Citation9 The cohesin protector Sgo1 also protects centriole cohesion.Citation10 Because cohesin defects cause chromosome missegregation, which can lead to abnormal spindle formation in animal cells,Citation11 whether cohesin contributes directly to centrosome duplication and separation is controversial.Citation12,Citation13
We have recently reported that the Aurora kinase Ipl1 in yeast is required for the maintenance of a tight association between duplicated sister SPBs, which we termed SPB cohesion.Citation14 Premature loss of SPB cohesion leads to the formation of supernumerary SPBs and multipolar spindles during yeast meiosis. In addition, the Polo-like kinase Cdc5 is antagonistic to Ipl1 in meiotic SPB regulation.Citation14 The opposing roles of Ipl1 and Cdc5 at the meiotic SPB are reminiscent of their roles in regulation of sister-chromatid cohesion during meiosis I, when Ipl1 protects centromeric cohesion,Citation15,Citation16 whereas Cdc5 promotes meiotic cohesin removal from the chromosome to disassociate sister chromatids.Citation17 To determine whether cohesin function is required for meiotic SPB regulation, we performed live-cell microscopy to observe the dynamics of Spc42-marked yeast SPBs in cohesin mutants (). Spc42 is a core component of the yeast SPB.Citation18 Both rec8Δ and PCLB2‑SCC3 (which depletes Scc3 in meiosis) mutant cells showed the formation of supernumerary Spc42‑GFP foci (), a result reminiscent of that in Ipl1‑depleted meiotic cells.Citation14 To determine whether stall of cell progression at prophase I combined with a defective meiotic recombination, which could occur in cohesin mutant cells, was responsible for formation of supernumerary Spc42 foci, we blocked the cells at prophase I with unrepaired double-strand breaks by deleting the DMC1 gene and observed essentially no formation of extra Spc42 foci in dmc1D cells (). In addition, as in Ipl1‑depleted cells, sister SPBs in cohesin mutants formed the doublet configuration due to the loss of SPB cohesion well before their complete separation in MI (), revealing that cohesin plays a role in SPB cohesion. In contrast to those of Ipl1‑depleted cells, however, the new Spc42 foci formed in the cohesin mutants failed to establish multipolar spindles, and the number of γ‑tubulin (called Tub4 in budding yeast) foci did not exceed four during meiosis in cohesin mutants as observed by fluorescence microscopy (our unpublished data), suggesting that only a portion of these Spc42‑containing bodies was capable of maturing into fully functional SPBs during yeast meiosis. Together, these data imply that cohesin is required for proper maintenance of SPB cohesion and support the conclusion that SPB cohesion leads to a restriction of SPB duplication,Citation14 but we cannot currently rule out the possibility that the supernumerary Spc42 foci formed in cohesin mutants during meiosis could represent the “dead poles” due to the overproduction of Spc42 as previously observed in vegetative yeast cells.Citation18
Our genetic investigation showed that cohesin is required for regulating SPB dynamics because it restricts the formation of extra Spc42‑containing bodies, revealing that cohesin plays a dual role in regulating sister-chromatid cohesion and SPB cohesion in yeast meiosis. Our findings support the conclusion from recent work with animal cells that cohesin plays an evolutionarily conserved role in centrosome cohesion and duplication. If cohesin directly mediates SPB cohesion and thus contributes to accurate SPB duplication at interphase II in yeast meiosis, we would expect cohesin to localize to the SPB, but our immuno-EM analysis of cohesin subunit Rec8 failed to localize cohesin to the SPB (our unpublished data). More importantly, meiotic SPBs were separated and formed a bipolar spindle in the presence of noncleavable cohesin, which served as the only source of cohesin in these cells during meiosis (). Our findings are therefore in contrast to those in a recent report that noncleavable Scc1 (homolog of Rec8) blocks centriole disengagement in animal tissue-culture cells.Citation12 Although we cannot currently rule out the possibility that cohesin localizes only transiently at the SPB to mediate SPB cohesion, we prefer a model in which cohesin contributes indirectly to SPB cohesion and duplication by ensuring proper chromosome segregation and spindle microtubule dynamics during yeast meiosis.
Coordination of chromosome segregation with centrosome dynamics is essential to ensuring genome integrity in all eukaryotes. In budding yeast, two cell-cycle-regulated kinases, the Aurora kinase Ipl1 and the Polo-like kinase Cdc5, play antagonistic roles in modulating sister-chromatid cohesion and SPB cohesion. Cohesin is a substrate of Cdc5 (and probably of Ipl1) and is required for topologically entrapping paired sister chromatids.Citation3 The requirement for cohesin in centrosome regulation is an exciting new development but is currently controversial. Future studies will clarify whether cohesin is directly involved in regulation of centrosome cohesion and duplication and, if so, how.
| Abbreviations: | ||
| SPB | = | spindle pole body |
| WT | = | wild type |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank K. Shirk for providing technical assistance. This work is supported in part by the National Science Foundation [MCB-0718384].
References
- Adams IR, Kilmartin JV. Spindle pole body duplication: a model for centrosome duplication?. Trends Cell Biol 2000; 10:329 - 35; http://dx.doi.org/10.1016/S0962-8924(00)01798-0; PMID: 10884685
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 2008; 24:105 - 29; http://dx.doi.org/10.1146/annurev.cellbio.24.110707.175350; PMID: 18616427
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet 2009; 43:525 - 58; http://dx.doi.org/10.1146/annurev-genet-102108-134233; PMID: 19886810
- Guan J, Ekwurtzel E, Kvist U, Yuan L. Cohesin protein SMC1 is a centrosomal protein. Biochem Biophys Res Commun 2008; 372:761 - 4; http://dx.doi.org/10.1016/j.bbrc.2008.05.120; PMID: 18515072
- Wong RW, Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc Natl Acad Sci USA 2008; 105:15441 - 5; http://dx.doi.org/10.1073/pnas.0807660105; PMID: 18832153
- Kong X, Ball AR Jr., Sonoda E, Feng J, Takeda S, Fukagawa T, et al. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol Biol Cell 2009; 20:1289 - 301; http://dx.doi.org/10.1091/mbc.E08-04-0419; PMID: 19116315
- Nakamura A, Arai H, Fujita N. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J Cell Biol 2009; 187:607 - 14; http://dx.doi.org/10.1083/jcb.200906019; PMID: 19948489
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 2006; 442:947 - 51; http://dx.doi.org/10.1038/nature04985; PMID: 16862117
- Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol 2007; 178:345 - 54; http://dx.doi.org/10.1083/jcb.200701163; PMID: 17664331
- Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell 2008; 14:331 - 41; http://dx.doi.org/10.1016/j.devcel.2007.12.007; PMID: 18331714
- Dai J, Kateneva AV, Higgins JM. Studies of haspin-depleted cells reveal that spindle-pole integrity in mitosis requires chromosome cohesion. J Cell Sci 2009; 122:4168 - 76; http://dx.doi.org/10.1242/jcs.054122; PMID: 19910498
- Schöckel L, Mockel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol 2011; 13:966 - 72; http://dx.doi.org/10.1038/ncb2280; PMID: 21743463
- Tsou MFB, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell 2009; 17:344 - 54; http://dx.doi.org/10.1016/j.devcel.2009.07.015; PMID: 19758559
- Shirk K, Jin H, Giddings TH Jr., Winey M, Yu H-G. The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J Cell Sci 2011; 124:2891 - 6; http://dx.doi.org/10.1242/jcs.086652; PMID: 21878496
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 2007; 128:477 - 90; http://dx.doi.org/10.1016/j.cell.2006.12.040; PMID: 17289568
- Yu H-G, Koshland DE. The Aurora kinase Ipl1 maintains the centromeric localization of PP2A to protect cohesin during meiosis. J Cell Biol 2007; 176:911 - 8; http://dx.doi.org/10.1083/jcb.200609153; PMID: 17371833
- Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 2003; 300:482 - 6; http://dx.doi.org/10.1126/science.1081846; PMID: 12663816
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol 2004; 20:1 - 28; http://dx.doi.org/10.1146/annurev.cellbio.20.022003.114106; PMID: 15473833
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 2000; 103:387 - 98; http://dx.doi.org/10.1016/S0092-8674(00)00131-8; PMID: 11081626