Abstract
Ribonucleoprotein-containing granules in the cytoplasm of germinal cells are known to be a common attribute of eukaryotic organisms. Germ granules appear to ensure the posttranscriptional regulation of germline mRNAs. Recent studies specify the participation of the germ granules in genome integrity maintenance by mechanisms involving short piRNAs. PIWI clade proteins and associated piRNAs are considered as key participants of the germline-specific piRNA pathway. Proteins of the PIWI clade, Aub and AGO3, concentrated in the germline-specific perinuclear granules called nuage, are involved in silencing of retrotransposons and other selfish repetitive elements in the Drosophila genome. In Drosophila testes, two types of perinuclear nuage granules are found: a large amount of small particles around the nuclei and significantly larger structures, the piNG-bodies. In this mini-review, we analyze the recent published data about structure and functions of Drosophila male germ granules, and especially their involvement in the piRNA silencing pathway.
Cytoplasmic RNA-rich non-membranous structures, which are remarkably conserved in germinal tissues of many eukaryotic species, are currently designated as germ granules. More than a century ago, a perinuclear granule named chromatiod body (CB) was found in the cytoplasm of mammalian male germ cells from late spermatocytes to round spermatids.Citation1 Germ granules differ significantly in their morphology and functions in various species, for example, polar granules or pole plasm in D. melanogaster, C. elegans, X. laevis, nuage and sponge body in D. melanogaster, P granules in C. elegans, CB and intermitochondrial cement (IMC or pi-body) in mammals.Citation1,Citation2 CB is thought to be cognate to the germ cell specification structure, nuage, in Drosophila. It has been proposed that germ granules contain proteins and mRNAs needed for germline development and gametogenesis. Germ granules also shared some protein components with cytoplasmic granules of somatic cells known as P-bodies or processing bodies.Citation3 However, their molecular functions remain mysterious to this day. Recent studies suggest the involvement of germ granules in defense of the germline genome from active endogenous elements, such as transposons.Citation4 Maintaining genome integrity during spermatogenesis and oogenesis is critical for a species viability. Discoveries of the past ten years revealed the essential role of gonad-specific piRNAs (small RNAs of 25–30 nt that associate with proteins of PIWI clade) in the silencing of transposons.Citation5-Citation10 This is now considered as a general mechanism of genome defense in the germlines of plants, fungi, worms, insects and mammals.Citation4 piRNAs are the largest and most complicated class of small RNAs. Biogenesis of piRNAs and piRNA-mediated posttranscriptional silencing appear to take place mainly in the cytoplasmic germ granules.Citation11-Citation14 In this mini-review we focus on the recent investigations of male germ granules from the well-known eukaryotic model organism, Drosophila, and their involvement to the piRNA silencing pathway.
Nuage as an Organelle Composed of Germ Granules
Perinuclear granules in germ cells of Drosophila have been named nuage (which means “cloud” in French). Nuage was described at first in nurse cells of the Drosophila ovaries.Citation11,Citation15-Citation17 These structures are visible as discontinuous rings around the nuclei on confocal slices. Similar structures were also detected in spermatogonial cells and primary spermatocytes in the testes.Citation17-Citation19 The main marker and essential component of nuage in both sexes is RNA-helicase Vasa.Citation16,Citation17,Citation20 Proteomic content of ovarian nuage granules includes: proteins of the PIWI subfamily, Aubergine and Argonaute 3; RNA-helicases, Vasa, Spindle E and Belle; Tudor domain-containing proteins, Tudor, Spindle E, Krimper, Tejas; proteins known as components of somatic P-bodies taking part in mRNA degradation, DCP1, Me31B and Pacman; and other proteins, such as putative nuclease Squash and high mobility box group protein Maelstrom.Citation11,Citation12,Citation15-Citation17,Citation21-Citation24 Nuage is thought to be involved in the selection and translational control of mRNAs transported from the nucleus.Citation12,Citation16,Citation17 Recent studies revealed the association of nuage with the piRNA biogenesis and piRNA-dependent silencing of transposons.Citation11,Citation12
Proteins of the PIWI subfamily, Piwi, Aubergine (Aub) and Argonaute 3 (AGO3), form complexes with piRNAs. These complexes recognize mRNAs complementary to guide piRNAs and perform RNA slicing, causing target degradation. PIWI clade proteins are considered the main players of the piRNA pathway.Citation5-Citation8,Citation10,Citation25 Two main models of piRNA processing have been suggested based on deep-sequencing analysis of ovarian piRNAs. In germinal cells of the ovaries, piRNAs complementary to transposon transcripts undergo amplification via the “ping-pong” cycle.Citation7,Citation8 Aub interacts mainly with antisense piRNAs derived from specialized genomic “master” loci, and AGO3 preferentially associates with the sense ones.Citation7,Citation8,Citation10,Citation25 Sense transposon transcripts are targeted by the Aub-containing antisense silencing complexes, which cause transcript cleavage. The cleavage act results in the production of 5′-termini of sense piRNAs. The details of 3′-processing of piRNAs are unknown up to now. These sense piRNAs are subsequently loaded into the AGO3-containing sense silencing complexes, which presumably recognize long antisense transcripts and generate 5′-ends of antisense piRNAs, which are involved in the next round of the cycle.Citation7,Citation8 The amplification cycle produces pairs of sense and antisense piRNAs which have a ten nucleotide overlap of complementary sequence in their 5′-ends. Nuage in nurse cells of the Drosophila ovaries is considered to be a site for both the piRNA maturation and the piRNA-guided silencing.Citation10-Citation12,Citation25 The third member of PIWI subfamily, nuclear protein Piwi, is expressed in both germline and somatic cells of the ovaries. In somatic cells in the absence of germline-specific Aub and AGO3 Piwi-associated piRNAs are produced by another mechanism, presumably via direct cleavage of long transcripts (primary processing). The ping-pong cycle is shown to be independent from Piwi.Citation7,Citation10 However, Piwi is essential for transposon silencing in the germline,Citation6,Citation10,Citation25 and, according to recent data, is involved in co-transcriptional repression of telomeric retrotransposons.Citation26
piRNA Pathway in the Testes
The first case of natural piRNA-mediated silencing of endogenous genes was discovered in the Drosophila testes.Citation5 However, the majority of papers published since then is devoted to the piRNA silencing in the ovaries, where powerful transposon activity is easily detectable in piRNA pathway mutant flies. Here we summarize the known peculiarities of the piRNA pathway in the testes.
Transcripts of the X-linked endogenous tandemly repeated Stellate genes, but not of transposons, are shown to be the main targets of the piRNA silencing in the testes of D. melanogaster.Citation18,Citation27 Stellate gene expression is strongly repressed by the piRNA machinery in wild type males. In case of a deletion of the homologous Y-linked crystal or Suppressor of Stellate (Su(Ste)) locus, abnormally high-level Stellate expression occurs in primary spermatocytes and leads to the accumulation of protein crystals, disturbances in meiotic chromosome condensation and segregation and essentially reduced fertility.Citation28,Citation29 Abundant anti-sense transcription of the crystal locus is responsible for the production of antisense Su(Ste) piRNAs complementary to Stellate transcripts. Nuage proteins, such as Aub, AGO3, Spindle E, Armitage, Tejas, Krimper, Maelstrom, Vasa and Squash are found to be necessary for the piRNAs-mediated Stellate silencing. Mutations in these genes result in the loss of Su(Ste) piRNAs and Stellate derepression.Citation5,Citation6,Citation18,Citation19,Citation24,Citation25,Citation30,Citation31 It should be noted that the proteins, which deficiencies lead to Stellate derepression, are largely concentrated to nuage. Piwi, the founder of PIWI clade, is known to be expressed mainly in somatic cells of the testis germinal proliferative center.Citation27 It is considered dispensable for Stellate silencing.Citation6
Large-scale sequencing of testis Aub-associated short RNAs revealed that 70% of these piRNAs were Su(Ste)-derived, predominately in antisense orientation.Citation18 About 90% of Su(Ste) piRNAs were presented by a piRNA of a single type, called Su(Ste)-4, or its variants. This observation strongly favors the assumption that Su(Ste) piRNAs are non randomly produced from precursor anti-sense Su(Ste) transcripts, but rather from a limited number of “hotspots.” However, among AGO3-associated piRNAs only 6% belongs to Su(Ste) piRNAs abundantly represented by Su(Ste)-4 too. Only a trace amount of sense Su(Ste) piRNAs was found in Aub and AGO3 complexes.Citation18 Also, a few complementary pairs of Su(Ste) piRNAs overlapping by 5′-ends (bearing signatures of the ping-pong cycle) were found. Thus, the bulk of testis piRNAs is generated by primary processing rather than the ping-pong mechanism. In spite of AGO3 expression level being significantly lower in the testes than that of Aub,Citation18 the biogenesis of Su(Ste) piRNAs and Stellate silencing are dependent on both Aub and AGO3.Citation5,Citation18,Citation25 A biological rationale for these requirements is still unclear now.
Only 54% of piRNAs from the AGO3-derived library and 7% from the Aub-derived one belong to transposon sequences.Citation18 A significant part of transposon-associated piRNAs demonstrated ping-pong signatures and generated ping-pong pairs similarly to the situation in the ovaries. However, contrary to their effects in the ovaries, Aub and AGO3 deficiencies only slightly affect transposon expression in the testes.Citation5,Citation18 Apparently, mobilization of transposons in the testes is essentially under the control of an alternative mechanism.
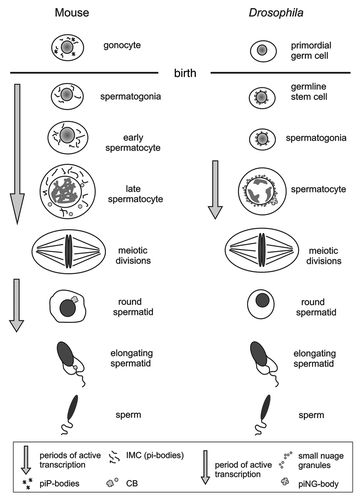
In our recent paperCitation19 we found at least two types of perinuclear nuage granules in germinal cells of the testes: a lot of small particles of about 0.6 µm that were concentrated around the nuclei, and significantly larger structures of about 2.4 µm, usually one per cell. The volume of larger granules is found to be more than 50 times that of the smaller ones. Large granules are enriched by the known nuage proteins: Vasa, Aub, AGO3, Tudor, Spindle E, Belle, Squash, Cutoff and also AGO1, the principal component of the microRNA pathway. Since most of the identified components are known as participants of the piRNA pathway, the new structure was named the piNG-body (piRNA Nuage Giant body). Vasa and Aub are shown to be completely colocalized at the periphery of the piNG-bodies, whereas AGO3 is located in the central lobe. The piNG-bodies appear in primary spermatocytes during active transcription period and are lost before meiotic divisions (). Neither piNG-bodies nor small nuage granules are detected in round spermatids. Mutational analysis demonstrated a strong association of piNG-bodies formation with the Stellate silencing.Citation19 Symmetrical arginine methylation of PIWI clade proteins provided by arginine methyltransferase Capsuleen (Dart5)Citation32 is found to be essential for piNG-body formation. Capsuleen mutations leading to Aub delocalization to random cytoplasmic foci induce strong Stellate derepression and loss of Su(Ste) piRNAs.Citation19 Since AGO1 is found to be a piNG-body component, the functions of the large granules in the microRNA-mediated mRNA silencing can also be expected. In addition to that we detected DCP1, a decapping enzyme and marker of somatic P-bodies, as a piNG-body constituent (Kibanov, unpublished data).
Figure 1. Scheme of spermatogenesis in mouse and Drosophila. Left: Germ granules in mouse gonocytes (IMC or pi-bodies, piP-bodiesCitation13,Citation14), spermatogonia, early sprematocytes (IMC), late spermatocytes (IMC, CB precursors) and haploid round spermatids (CB) are depicted. Adapted from ref. Citation2. CB formation coincides with periods of active transcription (arrows) and emerging of abundant pachytene piRNAs. In elongating spermatids the CBs gradually degenerates. Right: Two types of germ granules are found in the Drosophila male germline. There are a lot of small nuage granules, that appear early in germline stem cells, and significantly larger structures, the piNG-bodies. The piNG-bodies form in primary spermatocytes and disappear before meiotic divisions. Arrow indicates massive transcription during spermatocyte growth and maturation. No germ granules are found in round spermatids, where transcription program dramatically ceases.
Whereas no piNG-body-like large nuage structures are observed in the ovaries, a question concerning their role in the testes is raised. Taking into account that the conventional nuage granules in the ovarian nurse cells appear to be enough for functioning of the “ping-pong” piRNA silencing mechanism, the piNG-bodies seem to be dispensable for the amplification cycle. We know that Stellate transcription occurs in primary spermatocytesCitation30 and temporarily coincides with the piNG-body formation.Citation19 The analysis of Su(Ste) piRNAs derived from the testis libraries favors their generation by primary processing rather than by the “ping-pong” cycle.Citation18 The arranged internal structure of the piNG-body and a high concentration of piRNA pathway componentsCitation19 seem to contribute to effective kinetics of the Stellate silencing process. Abundant transcription of antisense Su(Ste) starts in spermatogonial cells, earlier than that of Stellate genes.Citation30 It is tempting to speculate that Su(Ste) piRNAs are produced before the start of Stellate transcription by a variant of primary processing. However, details of Stellate repression by the piRNA pathway and Su(Ste) piRNA biogenesis remain obscure to date.
Functional Relations Between piNG-body and CB
The mammalian CB is another large granular organelle involved in posttranscriptional mRNA processing in the testes. Although it appears in late pachytene spermatocytes and finally forms in round spermatids as a single structure per cell (), its persistence, similar to the piNG-body, coincides with the strong wave of transcription.Citation1,Citation2 Now more than 40 various proteins are identified as CB components.Citation1,Citation2 The CB contains MVH, the mammalian homolog of Drosophila Vasa protein; MIWI and MILI, PIWI clade proteins; Tudor domain-containing proteins; Dicer, the nuclease necessary for the microRNAs processing.Citation33,Citation34 MicroRNAs and microRNA pathway proteins were found in the CBs suggesting their role in microRNA-mediated expression regulation.Citation33 The CBs also contain proteins of the general mRNA degradation machinery.Citation1,Citation2,Citation33,Citation34 Biochemical isolation of mouse CBs reveals a high concentration of piRNAs, named late or pachytene piRNAs,Citation34 which do not demonstrate ping-pong signatures unlike pre-pachytene piRNAs that are expressed earlier in gonocytes.Citation9,Citation13 Pachytene piRNAs originate mainly from large non-repeated clusters located in non-annotated genome regions, but not from transposon-related sequences.Citation35-Citation37 Targets of pachytene piRNAs are largely unknown. Although the exact roles of the CB and pachytene piRNAs in mouse spermatogenesis remain to be determined, we suggest that the piNG-body and CB have shared attributes and can be functionally related structures.
The recent studies of the piRNA pathway using Drosophila testes provide important insights into the biological role of the germ granules. Similar characteristics and protein content of the CB and piNG-body allow using Drosophila as the model organism for further investigation of molecular principles of the germ granules organization and functioning.
| Abbreviations: | ||
| CB | = | chromatoid body |
| IMC | = | intermitochondrial cement |
| Su(Ste) | = | Suppressor of Stellate |
| piRNA | = | PIWI-interacting RNA |
Acknowledgments
This work was supported by the Russian Foundation for Basic Research grants #10–04–00535-a and #11–04–00017-a and by the Molecular and Cellular Biology program, RAS. We thank Anastasia Stolyarenko for help in manuscript preparation.
References
- Yokota S. Historical survey on chromatoid body research. Acta Histochem Cytochem 2008; 41:65 - 82; http://dx.doi.org/10.1267/ahc.08010; PMID: 18787638
- Meikar O, Da Ros M, Korhonen H, Kotaja N. Chromatoid body and small RNAs in male germ cells. Reproduction 2011; 142:195 - 209; http://dx.doi.org/10.1530/REP-11-0057; PMID: 21652638
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 2007; 8:9 - 22; http://dx.doi.org/10.1038/nrm2080; PMID: 17183357
- Saito K, Siomi MC. Small RNA-mediated quiescence of transposable elements in animals. Dev Cell 2010; 19:687 - 97; http://dx.doi.org/10.1016/j.devcel.2010.10.011; PMID: 21074719
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 2001; 11:1017 - 27; http://dx.doi.org/10.1016/S0960-9822(01)00299-8; PMID: 11470406
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006; 313:320 - 4; http://dx.doi.org/10.1126/science.1129333; PMID: 16809489
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089 - 103; http://dx.doi.org/10.1016/j.cell.2007.01.043; PMID: 17346786
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007; 315:1587 - 90; http://dx.doi.org/10.1126/science.1140494; PMID: 17322028
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31:785 - 99; http://dx.doi.org/10.1016/j.molcel.2008.09.003; PMID: 18922463
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009; 137:522 - 35; http://dx.doi.org/10.1016/j.cell.2009.03.040; PMID: 19395010
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A 2007; 104:6714 - 9; http://dx.doi.org/10.1073/pnas.0701920104; PMID: 17428915
- Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 2009; 186:333 - 42; http://dx.doi.org/10.1083/jcb.200904063; PMID: 19651888
- Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 2009; 5:e1000764; http://dx.doi.org/10.1371/journal.pgen.1000764; PMID: 20011505
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 2009; 17:775 - 87; http://dx.doi.org/10.1016/j.devcel.2009.10.012; PMID: 20059948
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 2001; 128:2823 - 32; PMID: 11526087
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 2003; 130:859 - 71; http://dx.doi.org/10.1242/dev.00310; PMID: 12538514
- Snee MJ, Macdonald PM. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci 2004; 117:2109 - 20; http://dx.doi.org/10.1242/jcs.01059; PMID: 15090597
- Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 2010; 16:2503 - 15; http://dx.doi.org/10.1261/rna.2270710; PMID: 20980675
- Kibanov MV, Egorova KS, Ryazansky SS, Sokolova OA, Kotov AA, Olenkina OM, et al. A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing. Mol Biol Cell 2011; 22:3410 - 9; http://dx.doi.org/10.1091/mbc.E11-02-0168; PMID: 21775629
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 1994; 120:1201 - 11; PMID: 8026330
- Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol 2005; 277:92 - 101; http://dx.doi.org/10.1016/j.ydbio.2004.09.009; PMID: 15572142
- Arkov AL, Wang JY, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development 2006; 133:4053 - 62; http://dx.doi.org/10.1242/dev.02572; PMID: 16971472
- Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 2007; 12:851 - 62; http://dx.doi.org/10.1016/j.devcel.2007.03.022; PMID: 17543859
- Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 2010; 20:724 - 30; http://dx.doi.org/10.1016/j.cub.2010.02.046; PMID: 20362446
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009; 137:509 - 21; http://dx.doi.org/10.1016/j.cell.2009.04.027; PMID: 19395009
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, et al. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res 2011; 39:8703 - 11; http://dx.doi.org/10.1093/nar/gkr552; PMID: 21764773
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 2007; 13:1911 - 22; http://dx.doi.org/10.1261/rna.744307; PMID: 17872506
- Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics 1984; 107:611 - 34; PMID: 6430749
- Hardy RW, Lindsley DL, Livak KJ, Lewis B, Siversten AL, Joslyn GL, et al. Cytogenetic analysis of a segment of the Y chromosome of Drosophila melanogaster. Genetics 1984; 107:591 - 610; PMID: 6430748
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol 2004; 24:6742 - 50; http://dx.doi.org/10.1128/MCB.24.15.6742-6750.2004; PMID: 15254241
- Kotelnikov RN, Klenov MS, Rozovsky YM, Olenina LV, Kibanov MV, Gvozdev VA. Peculiarities of piRNA-mediated post-transcriptional silencing of Stellate repeats in testes of Drosophila melanogaster. Nucleic Acids Res 2009; 37:3254 - 63; http://dx.doi.org/10.1093/nar/gkp167; PMID: 19321499
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 2009; 11:652 - 8; http://dx.doi.org/10.1038/ncb1872; PMID: 19377467
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A 2006; 103:2647 - 52; http://dx.doi.org/10.1073/pnas.0509333103; PMID: 16477042
- Meikar O, Da Ros M, Liljenbäck H, Toppari J, Kotaja N. Accumulation of piRNAs in the chromatoid bodies purified by a novel isolation protocol. Exp Cell Res 2010; 316:1567 - 75; http://dx.doi.org/10.1016/j.yexcr.2010.02.023; PMID: 20219458
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006; 442:203 - 7; PMID: 16751777
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 2006; 20:1709 - 14; http://dx.doi.org/10.1101/gad.1434406; PMID: 16766680
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 2007; 316:744 - 7; http://dx.doi.org/10.1126/science.1142612; PMID: 17446352