Abstract
N-acetylglucosamine (GlcNAc) has long been known to play important roles in cell surface structure. Recent studies are now revealing new functions for GlcNAc in cell signaling. Exposure to GlcNAc regulates virulence functions in the human fungal pathogen Candida albicans and in pathogenic bacteria. These signaling pathways sense exogenous GlcNAc and are distinct from the O-GlcNAc signaling pathways in mammalian cells in which increased levels of intracellular GlcNAc synthesis leads to post-translational modification of proteins by attachment of O-GlcNAc. The novel roles of GlcNAc in cell signaling will be the subject of this mini-review.
GlcNAc plays important structural roles at the surface of a wide range of cells from bacteria to humans. It is a component of bacterial cell wall peptidoglycan, fungal cell wall chitin and also glycosaminoglycans and other polymers on the surface of animal cells. In view of its widespread occurrence, it is not too surprising that GlcNAc has also been found to induce cell signaling in fungi and bacteria. These organisms are thought to respond to exogenous sources of GlcNAc, such as the material released during the extensive cell wall remodeling that occurs in growing fungi and bacteria. Animal cells and plants also use GlcNAc for cell signaling, but do so in a different manner. In these multicellular organisms, intracellular levels of UDP-GlcNAc are used as sensors of nutritional status that result in protein modification by attachment of O-GlcNAc. Studies on the role of O-GlcNAc in cell signaling have been the subject of several recent reviews.Citation1,Citation2 Therefore, we will focus this review on emerging data indicating the presence of novel signal pathways activated by exogenous GlcNAc in fungi and bacteria. Studies on these GlcNAc pathways are revealing new insights into intercellular and interspecies communication.
GlcNAc Signaling in C. albicans and Other Fungi
Addition of GlcNAc to the extracellular medium activates signal transduction pathways in the yeasts C. albicans,Citation3 Candida lusitaniae,Citation4 and Yarrowia lipolytica.Citation5 GlcNAc induces these species to switch from growing as budding cells to forming long filamentous hyphal cells. GlcNAc signaling has been best studied in the human fungal pathogen C. albicans, as the more commonly studied model yeasts Saccharomyces cerevisiae and Schizzosaccharomyces pombe lack the genes required to catabolize GlcNAc and do not appear to respond to this sugar. Genetic analysis of C. albicans indicates that GlcNAc activates two pathways. One pathway results in increased cAMP signaling, which then triggers hyphal morphogenesis and expression of virulence factors.Citation6-Citation8 GlcNAc activation of cAMP signaling also induces an epigenetic switch known as White-Opaque switching that results in altered morphological characteristics and gene expression in mating type homozygous forms of C. albicans.Citation9 The second pathway activated by GlcNAc, which is independent of cAMP signaling, results in increased expression of the genes needed to catabolize GlcNAc.Citation8,Citation10,Citation11
Initial attempts to determine whether GlcNAc has to be taken up by C. albicans to induce signaling led to conflicting conclusions.Citation3,Citation12 This was due in part to the lack of genetic approaches available at the time to study this diploid organism. However, the more recent discovery of the GlcNAc transporter (Ngt1) in a proteomics study of C. albicans plasma membrane proteins has provided an important new tool for analysis of GlcNAc signaling.Citation13 NGT1 was identified as a GlcNAc transporter because an ngt1Δ deletion mutant was very strongly impaired in GlcNAc uptake, and heterologous expression of NGT1 in S. cerevisiae conferred the ability to take up GlcNAc. Additional studies showed that Ngt1 was specific for GlcNAc and did not promote the transport of other related sugars. Although ngt1Δ cells were strongly defective in GlcNAc uptake, they could still import low levels of this sugar. Thus, it was very significant that the ngt1Δ mutant cells could still be stimulated to induce hyphal morphogenesis in the presence of 1,000-fold higher levels of GlcNAc than are required to induce the wild type. These results support the conclusion that Ngt1 facilitates uptake of GlcNAc, and that intracellular GlcNAc then induces signaling in C. albicans. Interestingly, NGT1 has also turned out to be a valuable new tool for studying the roles of GlcNAc in other organisms, since it is the first eukaryotic GlcNAc transporter gene to be identified.Citation14
Subsequent studies now indicate that GlcNAc metabolism is not necessary for its ability to induce signaling in C. albicans.Citation15 This was tested in part by mutating the HXK1, NAG1 and DAC1 genes needed to catabolize GlcNAc and use it for energy. These genes encode the enzymes needed for GlcNAc to be phosphorylated, deacetylated and then deaminated, resulting in its conversion to fructose-6-PO4. Significantly, a triple mutant lacking all three genes could still respond to GlcNAc to induce both the formation of hyphae and increased expression of NGT1.Citation15 Since phosphorylation of GlcNAc by Hxk1 is also required for cells to convert GlcNAc into UDP-GlcNAc for use in anabolic pathways, the ability of the triple mutant to be stimulated indicates that GlcNAc metabolism is not required for signaling.
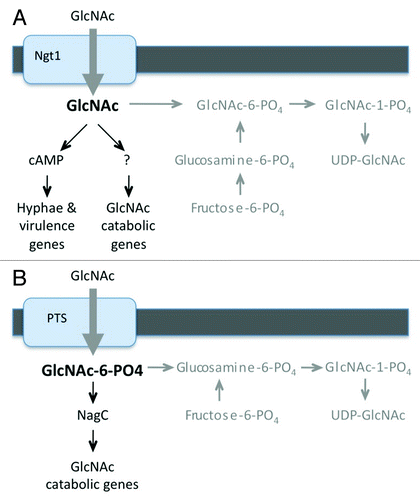
The ability of cells to induce signaling in the absence of the GlcNAc kinase (Hxk1) indicates that non-phosphorylated GlcNAc is capable of inducing C. albicans. This has important advantages for signaling as it allows cells to distinguish the non-phosphorylated GlcNAc transported into the cell from the GlcNAc-6-PO4 that is synthesized de novo within the cell (). GlcNAc synthesis involves conversion of fructose-6-PO4 to glucosamine-6-PO4 and then to GlcNAc-6-PO4.Citation16 Consequently, significant amounts of non-phosphorylated GlcNAc are not expected to occur in the cell unless it taken up from an exogenous source. Furthermore, detecting non-phosphorylated GlcNAc is also expected to provide a higher degree of sensitivity for low levels of extracellular GlcNAc imported into the cell in the presence of endogenously synthesized GlcNAc-6-PO4. Cells must synthesize large amounts of GlcNAc-6-PO4 to keep up with the demands for it in N-linked glycosylation, GPI anchor synthesis and synthesis of cell wall chitin. Not too surprisingly, signaling pathways activated by other sugars have also been reported to sense the non-phosphorylated form that is imported into the cell.Citation17,Citation18 Altogether, these results demonstrate that the capacity of C. albicans to sense non-phosphorylated GlcNAc taken up into the cell represents a novel signal transduction mechanism that may also occur in other organisms.
Figure 1. Models for GlcNAc induction of cell signaling. Note that (A) C. albicans and (B) bacteria such as E. coli are thought to sense forms of GlcNAc that are not synthesized in the cell. Endogenous GlcNAc synthesis pathways are shown in gray.
The response of C. albicans to GlcNAc raises the question of what might be the sources of GlcNAc that it could encounter. One possibility is that C. albicans responds to GlcNAc released by chitinases that remodel the cell wall during growth or released by the action of human chitinase during infection, since the fungal cell wall contains chitin, a polymer of GlcNAc. Similarly, GlcNAc is also released during growth of bacteria due to remodeling of the peptidoglycan cell wall layer, which is composed in part of GlcNAc.Citation19 This latter source of GlcNAc may be important for signaling in mixed microbial environments, such as the G.I. tract. GlcNAc is also an abundant component of the cell surface on mammalian cells, and is present in polymers such as glycosaminoglycans.Citation20
GlcNAc Signaling in Bacteria
GlcNAc has been shown to induce signaling in several different bacterial species. One example is Pseudomonas aeruginosa, which was recently reported to respond to GlcNAc in the lung secretions (sputum) of Cystic Fibrosis patients.Citation21 Microarray analysis revealed that growth in sputum induced expression of the GlcNAc catabolic genes in P. aeruginosa, consistent with the presence in sputum of GlcNAc-containing polymers, such as hyaluronic acid and mucins.Citation21 Interestingly, GlcNAc also induced the production of phenazine antimicrobial compounds by P. aeruginosa. It was proposed that this antimicrobial defense system might be induced by GlcNAc since P. aeruginosa also lives in the soil where GlcNAc would be an indicator of the presence of other bacteria or fungi in the area. Similarly, GlcNAc also induces production of antimicrobial compounds and regulates development during sporulation of some soil bacteria, such as Streptomyces coelicolor.Citation22
Another bacterial pathogen, Escherichia coli, responds to GlcNAc by reducing expression of CURLI fibers and type 1 fimbriae.Citation23,Citation24 CURLI fibers are important for biofilm formation, adhesion and the internalization of E. coli by epithelial cells.Citation23 Fimbriae are important for pathogenicity by promoting adhesion to mammalian cells.Citation24 Decreased production of these cell surface molecules is thought to promote dissemination of bacteria within the host. This response to GlcNAc may also be involved in balancing the interaction between E. coli and the host immune response, since CURLI and fimbriae are thought to be proinflammatory. GlcNAc may also have other roles in virulence, as suggested by studies showing that the ability of E. coli to catabolize GlcNAc is also important for it to colonize the G.I. tract.Citation25
GlcNAc also induces the genes needed for its catabolism in bacteria. In the bacteria where this has been studied, the GlcNAc catabolic genes are located in an operon that is regulated by a repressor, such as NagC in E. coli.Citation26 GlcNAc is taken up by bacteria through a phosphotransferase transporter system that results in GlcNAc-6-PO4 entering the cell. Binding of GlcNAc-6-PO4 to the NagC repressor causes allosteric changes that derepress the operon and promote expression of the genes needed for GlcNAc catabolism. Interestingly, bacteria are distinct from eukaryotic cells in that they do not synthesize GlcNAc-6-PO4.Citation16 They instead convert glucosamine-6-PO4 directly to GlcNAc-1-PO4. Intracellular GlcNAc-6-PO4is only expected to occur in bacteria as a result of the import of exogenous GlcNAc (). Thus, bacteria have a mechanism for distinguishing exogenous GlcNAc from the GlcNAc synthesized in the cell that is analogous to the detection of non-phosphorylated GlcNAc in C. albicans (). In this regard it is also interesting that GlcNAc-6-PO4 imported into Gram-positive bacteria acts as a cofactor for the ribozyme activity of the glmS mRNA, which results in its cleavage.Citation27 Since glmS encodes glucosamine-6-PO4 synthetase, the first committed step in the GlcNAc synthesis pathway, this specialized mechanism turns off GlcNAc synthesis in the presence of exogenous GlcNAc. Thus, there are at least two mechanisms in bacteria by which GlcNAc-6-PO4 regulates gene expression.
GlcNAc Signaling in Mammalian Cells and Plants
The ability of mammalian cells and plants to respond to exogenous GlcNAc has not been well studied. These organisms contain the genes needed to catabolize GlcNAc, so they presumably have mechanisms to regulate their expression. However, very little is known about other responses to exogenous GlcNAc. In contrast, there is a rapidly growing body of work demonstrating that plants and mammals respond to changes in the intracellular levels of GlcNAc. Changes in nutrition that result in elevated GlcNAc synthesis lead to increased formation of UDP-GlcNAc, which is a substrate for the enzyme O-GlcNAc transferase (OGT).Citation1 OGT modifies proteins by catalyzing the transfer of the GlcNAc moiety of UDP-GlcNAc to Ser or Thr residues on proteins. This is a reversible modification in animal cells analogous to protein phosphorylation, as they also encode an enzyme that removes O-GlcNAc from proteins. Many of the substrates that have been identified for OGT play key roles in cellular regulation, such as the transcription factors c-myc, p53 and NFκ-B.Citation2 This type of O-linked GlcNAc attachment has therefore been implicated in a range of human diseases, including cancer and diabetes.
Interspecies Communication
The presence of GlcNAc on the surface of so many different cell types makes it a good indicator of either the presence of other organisms or potentially an attack on the outer surface of a cell. Since cells in nature typically grow in mixed microbial environments, GlcNAc is therefore also well suited to be part of the communication that goes on between cells. An interesting example of this type of interspecies communication occurs between C. albicans and P. aeruginosa. GlcNAc and other bacterial cell wall breakdown products induce C. albicans to undergo hyphal morphogenesis.Citation3,Citation28 P. aeruginosa cells are then able to form a dense biofilm on the filamentous hyphal cells and kill them.Citation29 C. albicans also responds to a quorum factor produced by P. aeruginosa (3-oxo-C12 homoserinelactone) by restricting its growth to the budding pattern, thereby protecting it from being killed since budding cells are not attacked by P. aeruginosa.Citation30 Significantly, the P. aeruginosa cells are also on the receiving end of a signal from C. albicans. A quorum factor produced by C. albicans (farnesol) interferes with quorum signaling in P. aeruginosa and prevents induction of virulence factors.Citation31 Thus, GlcNAc is part of a complex exchange of signaling molecules between C. albicans and P. aeruginosa. Given the prevalence of GlcNAc, it seems likely to be involved in other forms of interspecies communication.Citation32,Citation33
Acknowledgments
Our research on GlcNAc signaling was supported by a Public Health Service grant awarded to J.B.K. from the National Institutes of Health (RO1 GM087368). S.N. was supported by a Training Grant (NIH T32CA009176) from the National Cancer Institute and S.P. was supported by a Training Grant (NIH T32AI007539) from the National Institute of Allergy and Infectious Diseasesof the National Institutes of Health.
References
- Slawson C, Copeland RJ, Hart GW. O-GlcNAc signaling: a metabolic link between diabetes and cancer?. Trends Biochem Sci 2010; 35:547 - 55; http://dx.doi.org/10.1016/j.tibs.2010.04.005; PMID: 20466550
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem 2011; 80:825 - 58; http://dx.doi.org/10.1146/annurev-biochem-060608-102511; PMID: 21391816
- Simonetti N, Strippoli V, Cassone A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature 1974; 250:344 - 6; http://dx.doi.org/10.1038/250344a0; PMID: 4605454
- Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 2009; 19:891 - 9; http://dx.doi.org/10.1016/j.cub.2009.04.058; PMID: 19446455
- Pérez-Campo FM, Domínguez A. Factors affecting the morphogenetic switch in Yarrowia lipolytica. Curr Microbiol 2001; 43:429 - 33; http://dx.doi.org/10.1007/s002840010333; PMID: 11685511
- Castilla R, Passeron S, Cantore ML. N-acetyl-D-glucosamine induces germination in Candida albicans through a mechanism sensitive to inhibitors of cAMP-dependent protein kinase. Cell Signal 1998; 10:713 - 9; http://dx.doi.org/10.1016/S0898-6568(98)00015-1; PMID: 9884022
- Leberer E, Harcus D, Dignard D, Johnson L, Ushinsky S, Thomas DY, et al. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol Microbiol 2001; 42:673 - 87; http://dx.doi.org/10.1046/j.1365-2958.2001.02672.x; PMID: 11722734
- Gunasekera A, Alvarez FJ, Douglas LM, Wang HX, Rosebrock AP, Konopka JB. Identification of GIG1, a GlcNAc-induced gene in Candida albicans needed for normal sensitivity to the chitin synthase inhibitor nikkomycin Z. Eukaryot Cell 2010; 9:1476 - 83; http://dx.doi.org/10.1128/EC.00178-10; PMID: 20675577
- Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog 2010; 6:e1000806; http://dx.doi.org/10.1371/journal.ppat.1000806; PMID: 20300604
- Kumar MJ, Jamaluddin MS, Natarajan K, Kaur D, Datta A. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc Natl Acad Sci U S A 2000; 97:14218 - 23; http://dx.doi.org/10.1073/pnas.250452997; PMID: 11114181
- Yamada-Okabe T, Sakamori Y, Mio T, Yamada-Okabe H. Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur J Biochem 2001; 268:2498 - 505; http://dx.doi.org/10.1046/j.1432-1327.2001.02135.x; PMID: 11298769
- Shepherd MG, Sullivan PA. The control of morphogenesis in Candida albicans. J Dent Res 1984; 63:435 - 40; http://dx.doi.org/10.1177/00220345840630031501; PMID: 6366000
- Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol Biol Cell 2007; 18:965 - 75; http://dx.doi.org/10.1091/mbc.E06-10-0931; PMID: 17192409
- Breidenbach MA, Gallagher JE, King DS, Smart BP, Wu P, Bertozzi CR. Targeted metabolic labeling of yeast N-glycans with unnatural sugars. Proc Natl Acad Sci U S A 2010; 107:3988 - 93; http://dx.doi.org/10.1073/pnas.0911247107; PMID: 20142501
- Naseem S, Gunasekera A, Araya E, Konopka JB. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J Biol Chem 2011; 286:28671 - 80; http://dx.doi.org/10.1074/jbc.M111.249854; PMID: 21700702
- Milewski S, Gabriel I, Olchowy J. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 2006; 23:1 - 14; http://dx.doi.org/10.1002/yea.1337; PMID: 16408321
- Sellick CA, Reece RJ. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem Sci 2005; 30:405 - 12; http://dx.doi.org/10.1016/j.tibs.2005.05.007; PMID: 15950477
- Gancedo JM. The early steps of glucose signalling in yeast. FEMS Microbiol Rev 2008; 32:673 - 704; http://dx.doi.org/10.1111/j.1574-6976.2008.00117.x; PMID: 18559076
- Doyle RJ, Chaloupka J, Vinter V. Turnover of cell walls in microorganisms. Microbiol Rev 1988; 52:554 - 67; PMID: 3070324
- Moussian B. The role of GlcNAc in formation and function of extracellular matrices. Comp Biochem Physiol B Biochem Mol Biol 2008; 149:215 - 26; http://dx.doi.org/10.1016/j.cbpb.2007.10.009; PMID: 18032081
- Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol 2011; 193:909 - 17; http://dx.doi.org/10.1128/JB.01175-10; PMID: 21169497
- Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Müller M, et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol 2006; 61:1237 - 51; http://dx.doi.org/10.1111/j.1365-2958.2006.05319.x; PMID: 16925557
- Barnhart MM, Lynem J, Chapman MR. GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J Bacteriol 2006; 188:5212 - 9; http://dx.doi.org/10.1128/JB.00234-06; PMID: 16816193
- Sohanpal BK, El-Labany S, Lahooti M, Plumbridge JA, Blomfield IC. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc Natl Acad Sci U S A 2004; 101:16322 - 7; http://dx.doi.org/10.1073/pnas.0405821101; PMID: 15534208
- Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 2004; 101:7427 - 32; http://dx.doi.org/10.1073/pnas.0307888101; PMID: 15123798
- Plumbridge JA. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol Microbiol 1991; 5:2053 - 62; http://dx.doi.org/10.1111/j.1365-2958.1991.tb00828.x; PMID: 1766379
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004; 428:281 - 6; http://dx.doi.org/10.1038/nature02362; PMID: 15029187
- Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H, et al. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 2008; 4:28 - 39; http://dx.doi.org/10.1016/j.chom.2008.05.014; PMID: 18621008
- Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 2002; 296:2229 - 32; http://dx.doi.org/10.1126/science.1070784; PMID: 12077418
- Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 2004; 54:1212 - 23; http://dx.doi.org/10.1111/j.1365-2958.2004.04349.x; PMID: 15554963
- Cugini C, Calfee MW, Farrow JM 3rd, Morales DK, Pesci EC, Hogan DA. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol 2007; 65:896 - 906; http://dx.doi.org/10.1111/j.1365-2958.2007.05840.x; PMID: 17640272
- Jarosz LM, Ovchinnikova ES, Meijler MM, Krom BP. Microbial spy games and host response: roles of a Pseudomonas aeruginosa small molecule in communication with other species. PLoS Pathog 2011; 7:e1002312; http://dx.doi.org/10.1371/journal.ppat.1002312; PMID: 22114549
- Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol 2010; 8:340 - 9; http://dx.doi.org/10.1038/nrmicro2313; PMID: 20348933