Abstract
Cytoplasmic poly(A)-binding proteins (PABPs) regulate mRNA stability and translation. Although predominantly localized in the cytoplasm, PABP proteins also cycle through the nucleus. Recent work has established that their steady-state localization can be altered by cellular stresses such as ultraviolet (UV) radiation, and infection by several viruses, resulting in nuclear accumulation of PABPs. Here, we present further evidence that their interaction with and release from mRNA and translation complexes are important in determining their sub-cellular distribution and propose an integrated model for regulated nucleo-cytoplasmic transport of PABPs.
Poly(A)-binding protein 1 (PABP1) is the prototypical member of the cytoplasmic PABP family and plays key roles in the regulation of mRNA translation and stability. By binding the 3′ poly(A) tail and interacting with translation factors [eukaryotic initiation factor 4G (eIF4G) and PABP-interacting protein-1 (PAIP1)] bound to the 5' end of mRNAs, PABP1 brings about a “closed-loop” mRNA conformation which promotes translation initiation by enhancing ribosome recruitment.Citation1,Citation2 This conformation also prevents mRNA deadenylation and decay, although PABP1 also paradoxically plays a role in the recruitment of deadenylase complexes.Citation1-Citation3 PABP1 has other roles in mRNA-specific translational regulationCitation4 and in preventing non-sense mediated decay in the absence of premature termination codons.Citation5,Citation6 Mammals encode three additional PABP proteins which share a common domain organization with PABP1: PABP4 which appears to be widely expressedCitation7 and testes PABP and embryonic PABP, which in adults appear largely restricted to the gonads.Citation8 Available evidence suggests that the ability to bind RNA and to participate in translational activation may be conserved across the family.Citation9-Citation13
Unsurprisingly, given the central role of PABP1 in orchestrating gene expression, both its levels and activity are finely regulated.Citation14-Citation16 PABP1 activity is controlled by PABP-interacting protein 2 (PAIP2) which effectively sequesters PABP1,Citation16 and recent evidence suggests that numerous post-translational modifications may also play a role in coordinating its various functions.Citation17 The sub-cellular localization of PABP1 is also subject to regulation.
PABP1 is a nucleo-cytoplasmic shuttling protein whose steady-state localization is predominantly cytoplasmic.Citation18 Interestingly, a number of cellular stresses result in PABP1 relocalization to stress granules (SGs) whereas infection by several viruses or treatment with transcriptional inhibitors cause PABP1 redistribution to the nucleus.Citation18-Citation23 Intriguingly, while UV treatment induces stress granules in a minority of cells at early time points after exposure, PABP1 is robustly relocalized to the nucleus at later times in both mouse and human cell lines.Citation19 Endogenous PABP4 shows a similar distribution to PABP1 in both normally growing and UV-stressed cells, although its kinetics of redistribution to the nucleus appear slower.Citation19
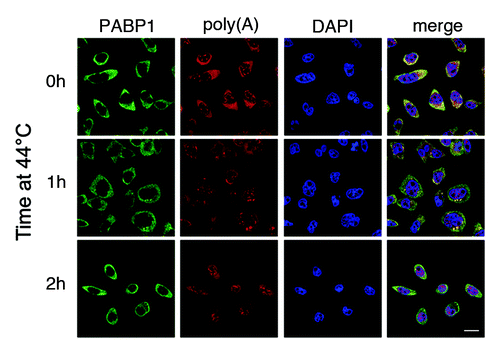
UV-induced nuclear relocalization of PABPs mirrors a change in the distribution of polyadenylated RNA, which accumulates in the nucleus indicative of a block in mRNA export.Citation19 Since stress granules are also foci for poly(A) RNA, this led us to hypothesize that mRNA distribution may be a major regulator of PABP localization.Citation19 In keeping with this idea, RNA changes accompanied PABP redistribution in cells ectopically expressing the herpes simplex virus-1 (HSV-1) protein ICP27 or treated with the transcriptional inhibitor actinomycin D, both of which indirectly inhibit mRNA export.Citation19 Importantly, direct inhibition of mRNA export by RNAi knockdown of the bulk mRNA export adaptor TAP resulted in nuclear relocalization of PABPs, demonstrating that nuclear export of PABP1 and PABP4 is dependent on mRNA export.Citation19 Other cellular stresses have been reported to cause nuclear relocalization of PABP1 including prolonged (2 h) heat shock,Citation24 a perturbation normally associated with stress granule formation. However, we observe neither accumulation of PABPs nor poly(A) RNA in the nucleus under prolonged heat shock (), suggesting this heat shock response may only occur in a subset of HeLa cells and in keeping with a model in which mRNA export plays an important role in determining PABP localization.Citation19
Figure 1. Heat shock results in PABP1 relocalization to stress granules but not the nucleus. HeLa cells were incubated at 44°C for the indicated time and fixed. Poly(A)-RNA (red) and PABP1 (green) were detected by FISH using an oligo dT40 probe and immunofluorescence, respectively.Citation19 Stress granules are visible after 1 h treatment as cytoplasmic foci containing both PABP1 and poly(A) RNA. Scale bars: 20 µm.
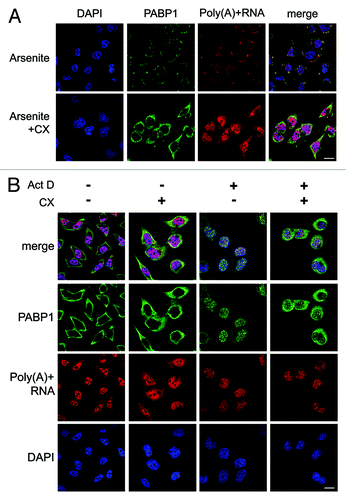
In unstressed cells PABP1 is undetectable by immunofluorescence in the nucleus implying that while PABP nuclear import and export are on-going only a small fraction of total PABP cycles through the nucleus at any given time with the majority being retained in the cytoplasm. Consistent with this, nuclear accumulation of PABPs requires several hours of transcriptional inhibition.Citation19 To test whether release from translation complexes in the cytoplasm is first required for the nuclear import of PABPs by α-importins,Citation25 we pre-treated cells with cycloheximide. This stabilizes polysomes (mRNAs being translated by multiple ribosomes) by inhibiting translation elongation. Cycloheximide treatment was shown to be effective since the formation of stress granules, which require polysome disassembly, was abrogated (). Actinomycin D was used inhibit transcription and induce nuclear relocalization of PABP1 and, in the absence of cycloheximide, relocalization was robust with very little PABP1 remaining in the cytoplasm (). Pre-treatment with cycloheximide had a significant impact on relocalization with a substantial portion of PABP1 remaining cytoplasmic, suggesting that release of PABP from polysomes is necessary for its nuclear import. This is consistent with findings that nuclear relocalization of PABP1 during rotavirus infection is dependent on its eviction from translation complexes by the viral protein NSP3, which competes with PABP for eIF4G binding.Citation26 Furthermore, recent work from the Glaunsinger lab has elegantly shown that the interaction of PABP1 with RNA can inhibit its interaction with α-importins.Citation25 Interestingly in this regard, cytoplasmic mRNA levels decrease in UV-treated cellsCitation19 and a modest increase in levels of cytoplasmic poly(A) RNA was noted in actinomycin D treated cells after cycloheximide pre-treatment ().
Figure 2. Pre-treatment with cycloheximide limits nuclear relocalization of PABP1. HeLa cells were pretreated with cycloheximide (CX, 100 μg/ml) for 30 min prior to and during treatment with (A) sodium arsenite (0.5 mM; 1 h) or (B) actinomycin D (Act D, 5 μg/ml; 12 h). Appropriate vehicle control was applied to non-treated cells. Poly(A)-RNA (red) and PABP1 (green) were detected by oligo dT40 FISH and immunofluorescence.Citation19 Scale bars: 20 µm.
Together these data support a model in which RNA is a major determinant of the transport of PABP to and from the cytoplasm (). In normally proliferating mammalian cells a significant portion of PABP1, and here we establish also PABP4 (), is associated with actively translating mRNAs on polysomes and is thus unavailable for nuclear shuttling. Unbound PABP molecules are available for association with α-importins and can be imported to the nucleus. When in the nucleus, PABP1 has been shown to bind pre-mRNAs alongside the predominantly nuclear and functionally distinct poly(A)-binding protein, PABPN1, which binds nascent poly(A)-tails as they are added.Citation27 Therefore exported mRNAs likely leave the nucleus with a mixed population of poly(A)-binding proteins, comprised of a majority of PABPN1 and a minority of cytoplasmic PABPs, coating their poly(A)-tails. The latter do not contribute to mRNA exportCitation19 but may potentially prime mRNAs for efficient translation. Once the mRNA reaches the cytoplasm PABPN1 is exchanged for cytoplasmic PABPs in a translation-dependent process.Citation27 Thus, perturbations of the balance between PABP import and export, such as the introduction of viral endonucleases that destroy cytoplasmic mRNAsCitation28 or the cessation of mRNA exportCitation19 result in a redistribution of predominantly cytoplasmic PABPs to the nucleus.
Figure 3. A model for PABP nucleo-cytoplasmic transport. In normally growing cells most molecules of PABP1 and PABP4 are located within the cytoplasm. PABP nuclear import is dependent on interaction with α-importins. This can be retarded by PABP binding to RNA and participation in translational complexes, and may also be affected by interaction with other PABP partner proteins. In the nucleus PABP1 and PABP4 bind mRNAs alongside PABPN1. PABP1 and PABP4 do not contribute to mRNA export but rather are exported in an mRNA-dependent manner. Enhancing the levels of PABP1 available for import or blocking mRNA export results in an accumulation of PABP1 in the nucleus.
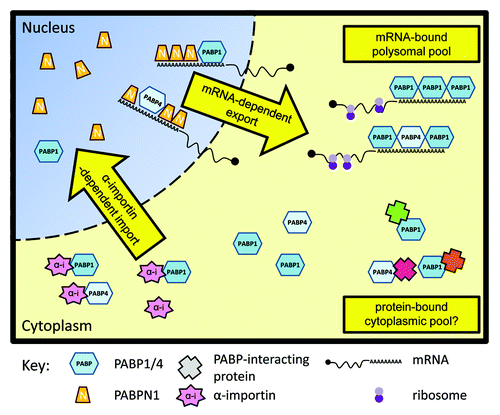
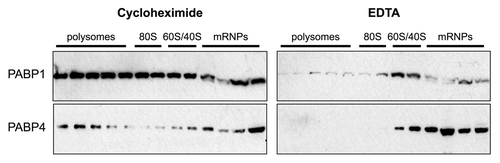
Figure 4. PABP1 and PABP4 are present in both polysomal and non-polysomal fractions. Post-nuclear HeLa extracts were sedimented through 10–50% sucrose gradients and fractionated. Proteins from the fractions were TCA precipitated prior to western blotting for PABP1 and PABP4. Cycloheximide (150 μg/ml) or EDTA (20 mM) was added to the lysis buffer to stabilize or disassemble polysomes respectively. Absorbance traces at 254nm were used to identify polysome, 80S, 60/40S and mRNP-containing fractions.Citation17
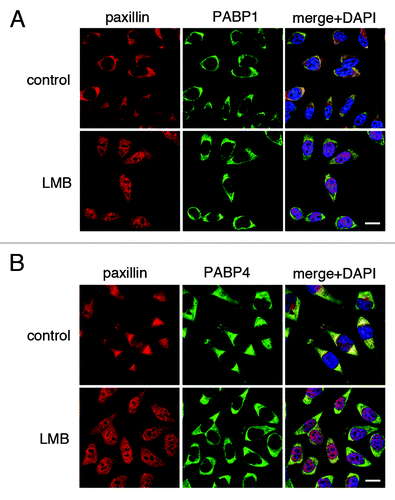
Changes in mRNA export or levels of cytoplasmic poly(A) RNA are likely to underlie the nuclear accumulation of PABP in many circumstances (e.g., following UV exposure) but may not be universally applicable, or may act in concert with additional mechanisms. For example, Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) encodes a highly abundant viral non-coding RNA, PAN (Polyadenylated nuclear RNA), which remains nuclear throughout infection and is proposed to sequester PABP1 within the nucleus.Citation29 Moreover, two non mRNA-dependent export pathways for mammalian PABPs have been described based on interactions with the translation elongation factor eEF1α or paxillin, although the extent to which these operate in parallel with mRNA-dependent export or are restricted to specific cell types is unclear.Citation30,Citation31 In murine 3T3 cells, where nuclear accumulation of PABP is accompanied by a block in mRNA-export after UV exposure,Citation19 nuclear relocalization of PABP1 has also been reported following inhibition of the CRM1 export pathway by leptomycin B.Citation32 This was attributed to an indirect effect of inhibiting nuclear export of paxillin, a protein best known for its role as a signaling adaptor molecule in cell adhesion complexes.Citation30 However, neither we nor Kumar et al. (2011)Citation25 could detect PABP-paxillin interactions in HEK293T or HeLa cells respectively (data not shown). Moreover, leptomycin B treatment of HeLa cells does not alter the localization of PABP1 or PABP4, despite significant relocalization of paxillin to the nucleus, indicating that paxillin export is effectively inhibited (). Taken together, these results suggest that PABP export mediated via proteins, such as paxillin, may only be significant within specific cell types. Identification of physiological conditions that disrupt paxillin or eEF1α-mediated PABP export may clarify their roles in directing PABP distribution.
Figure 5. Nuclear export of PABP1 and PABP4 is not prevented by the CRM1 inhibitor leptomycin B. HeLa cells were treated with 5 ng/ml leptomycin B (LMB) or vehicle control for 3 h prior to fixation and immunofluorescence detection of paxillin (red) and (A) PABP1 or (B) PABP4 (green).Citation19 Scale bars: 20 µm.
While release from polysomes () and RNACitation25 appears to be a pre-requisite for nuclear import of PABP1 and PABP4, in normally growing cells only a portion of these proteins is polysome associated (). In fact, PABP1 is present in 3-fold excess relative to the binding sites available on poly(A)-tails in HeLa cellsCitation33 and given that PABP4 is present at a ratio of 1:5 PABP1 molecules,Citation19 the true excess of PABP proteins over poly(A) in these cells is likely greater. However, despite the relative stoichiometry of PABP proteins to mRNA, PABPs do not flood the nucleus of unstressed cells unless overexpressed,Citation18 indicating that further mechanisms may exist to antagonize nuclear entry (). Protein partners may serve as additional cytoplasmic anchors by blocking interaction with α-importins, analogous to RNA. Indeed, several protein partners including regulatory proteins (e.g., PAIP2) as well as translation factors (eIF4G, PAIP1) interact with the RNA-recognition motif region of PABP1Citation4 that binds α-importin.Citation25 However, the relative abundance of these factors and whether they are able to compete with α-importins for PABP-binding remains to be established as does the binding sites for many other PABP protein partners.
Very little is known about the consequences of relocating cytoplasmic PABPs to the nucleus, or why PABPs are relocalized to the nucleus rather than stress granules during specific cellular stress responses. Nonetheless, it is tempting to speculate that movement of PABP to the nucleus vs. stress granules may serve as part of a complex reprogramming of protein synthesis required for cellular recovery in response to different cellular insults. Indeed, both relocalization of PABPs to the nucleus after UV exposure and reduction of cytoplasmic PABP levels by siRNA knockdown are associated with a modest decrease in global protein synthesis.Citation19 Since changes in the available levels of basal translation factors eIF4E and eIF4G alter the translational efficiency of certain mRNAs disproportionately,Citation34,Citation35 the partial removal of PABP from the cytoplasm may serve to reprogram translation by more profoundly effecting subsets of mRNAs. However, the extent to which PABP relocalization contributes to protein synthesis changes following UV-exposure remains to be determined since transcription and mRNA export are also altered. Changes in the levels of cytoplasmic PABP may also have effects on mRNA stability, although siRNA knockdown of PABPs did not result in obvious changes to overall cytoplasmic poly(A)-RNA levels examined by fluorescence in situ hybridization (FISH).Citation19 However, more subtle mRNA-specific effects would not have been detected and additional studies are required to understand the impact of PABP relocalization on gene expression within the cytoplasm.
The presence of PABPs in the nucleus raises questions pertaining to its function in this compartment. PABP1 and PABP4 are not required for mRNA export,Citation19 nor is their presence in the nucleus required for recovery from UV exposure, suggesting that their exclusion from the cytoplasm rather than import is important in recovery from UV exposure.Citation19 However, Kumar and Glaunsinger (2010)Citation36 found that accumulation of PABP1 in the nucleus results in aberrant hyperadenylation of transcripts. This hyperadenylation may be a result of increased levels of cytoplasmic PABPs in the nucleus interfering with the determination of poly(A) tail length by PABPN1.Citation37 However, FISH suggests that hyperadenylation may not occur following UV-exposure, a difference that may be due to the downregulation of transcription since polyadenylation occurs co-transcriptionally.Citation19 Furthermore, it is clear that hyperadenylation does not contribute to the block in mRNA export observed in UV-treated cells as this occurs before the majority of PABP accumulates in the nucleus and is affected by PABP1 and PABP4-knockdown.Citation19
Recent advances have clarified the picture of how PABP localization is regulated and underlined the important role of RNA. However, key questions clearly remain regarding the relative contribution and integration of different pathways and their regulation in response to physiological stimuli. Thus, as more factors emerge that alter PABP localization, it is increasingly important to understand the ins and outs of this mode of regulation and the cellular consequences of such changes.
| Abbreviations: | ||
| PABP | = | poly(A)-binding protein |
| poly(A) RNA | = | polyadenylated RNA |
| siRNA | = | short interfering RNA |
| PAN | = | polyadenylated nuclear RNA |
| KSHV | = | Kaposi’s sarcoma-associated herpes virus |
| UV | = | ultraviolet |
| FISH | = | fluorescence in situ hybridization |
| HSV-1 | = | herpes simplex virus-1 |
Acknowledgments
H.M.B. and N.K.G. were supported by MRC Unit funding (HGU/HRSU), an MRC Senior Non-Clinical Fellowship G117/564 and MRC transition funding to N.K.G. N.K.G. is also supported by an MRC program grant and BBSRC and Wellcome Trust project grants.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Gorgoni B, Gray NK. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct Genomic Proteomic 2004; 3:125 - 41; http://dx.doi.org/10.1093/bfgp/3.2.125; PMID: 15355595
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 2003; 4:223; http://dx.doi.org/10.1186/gb-2003-4-7-223; PMID: 12844354
- Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2011; 2:167 - 83; http://dx.doi.org/10.1002/wrna.40; PMID: 21957004
- Burgess HM, Gray NK. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem Soc Trans 2010; 38:1517 - 22; http://dx.doi.org/10.1042/BST0381517; PMID: 21118118
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 2007; 26:1591 - 601; http://dx.doi.org/10.1038/sj.emboj.7601588; PMID: 17318186
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 2008; 27:736 - 47; http://dx.doi.org/10.1038/emboj.2008.17; PMID: 18256688
- Yang H, Duckett CS, Lindsten T. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol 1995; 15:6770 - 6; PMID: 8524242
- Brook M, Smith JW, Gray NK. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction 2009; 137:595 - 617; http://dx.doi.org/10.1530/REP-08-0524; PMID: 19225045
- Sladic RT, Lagnado CA, Bagley CJ, Goodall GJ. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur J Biochem 2004; 271:450 - 7; http://dx.doi.org/10.1046/j.1432-1033.2003.03945.x; PMID: 14717712
- Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N, Steitz JA. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci U S A 2005; 102:367 - 72; http://dx.doi.org/10.1073/pnas.0408378102; PMID: 15630085
- Wilkie GS, Gautier P, Lawson D, Gray NK. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol Cell Biol 2005; 25:2060 - 71; http://dx.doi.org/10.1128/MCB.25.5.2060-2071.2005; PMID: 15713657
- Gorgoni B, Richardson WA, Burgess HM, Anderson RC, Wilkie GS, Gautier P, et al. Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc Natl Acad Sci U S A 2011; 108:7844 - 9; http://dx.doi.org/10.1073/pnas.1017664108; PMID: 21518916
- Kimura M, Ishida K, Kashiwabara S, Baba T. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol Reprod 2009; 80:545 - 54; http://dx.doi.org/10.1095/biolreprod.108.072553; PMID: 19020299
- de Melo Neto OP, Standart N, Martins de Sa C. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res 1995; 23:2198 - 205; http://dx.doi.org/10.1093/nar/23.12.2198; PMID: 7610048
- Hornstein E, Git A, Braunstein I, Avni D, Meyuhas O. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J Biol Chem 1999; 274:1708 - 14; http://dx.doi.org/10.1074/jbc.274.3.1708; PMID: 9880551
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, et al. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 2001; 7:205 - 16; http://dx.doi.org/10.1016/S1097-2765(01)00168-X; PMID: 11172725
- Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK. The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J 2012; 441:803 - 12; http://dx.doi.org/10.1042/BJ20111474; PMID: 22004688
- Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem 1998; 273:13015 - 21; http://dx.doi.org/10.1074/jbc.273.21.13015; PMID: 9582337
- Burgess HM, Richardson WA, Anderson RC, Salaun C, Graham SV, Gray NK. Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs. J Cell Sci 2011; 124:3344 - 55; http://dx.doi.org/10.1242/jcs.087692; PMID: 21940797
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 1999; 147:1431 - 42; http://dx.doi.org/10.1083/jcb.147.7.1431; PMID: 10613902
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 2010; 29:2841 - 57; http://dx.doi.org/10.1038/emboj.2010.143; PMID: 20606625
- Salaun C, MacDonald AI, Larralde O, Howard L, Lochtie K, Burgess HM, et al. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J Virol 2010; 84:8539 - 48; http://dx.doi.org/10.1128/JVI.00668-10; PMID: 20573819
- Smith RW, Gray NK. Poly(A)-binding protein (PABP): a common viral target. Biochem J 2010; 426:1 - 12; http://dx.doi.org/10.1042/BJ20091571; PMID: 20102337
- Ma S, Bhattacharjee RB, Bag J. Expression of poly(A)-binding protein is upregulated during recovery from heat shock in HeLa cells. FEBS J 2009; 276:552 - 70; http://dx.doi.org/10.1111/j.1742-4658.2008.06803.x; PMID: 19087191
- Kumar GR, Shum L, Glaunsinger BA. Importin alpha-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol Cell Biol 2011; 31:3113 - 25; http://dx.doi.org/10.1128/MCB.05402-11; PMID: 21646427
- Harb M, Becker MM, Vitour D, Baron CH, Vende P, Brown SC, et al. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J Virol 2008; 82:11283 - 93; http://dx.doi.org/10.1128/JVI.00872-08; PMID: 18799579
- Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol 2006; 26:3085 - 97; http://dx.doi.org/10.1128/MCB.26.8.3085-3097.2006; PMID: 16581783
- Lee YJ, Glaunsinger BA. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 2009; 7:e1000107; http://dx.doi.org/10.1371/journal.pbio.1000107; PMID: 19468299
- Borah S, Darricarrère N, Darnell A, Myoung J, Steitz JA. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 2011; 7:e1002300; http://dx.doi.org/10.1371/journal.ppat.1002300; PMID: 22022268
- Woods AJ, Kantidakis T, Sabe H, Critchley DR, Norman JC. Interaction of paxillin with poly(A)-binding protein 1 and its role in focal adhesion turnover and cell migration. Mol Cell Biol 2005; 25:3763 - 73; http://dx.doi.org/10.1128/MCB.25.9.3763-3773.2005; PMID: 15831480
- Khacho M, Mekhail K, Pilon-Larose K, Pause A, Côté J, Lee S. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol Biol Cell 2008; 19:5296 - 308; http://dx.doi.org/10.1091/mbc.E08-06-0562; PMID: 18799616
- Woods AJ, Roberts MS, Choudhary J, Barry ST, Mazaki Y, Sabe H, et al. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J Biol Chem 2002; 277:6428 - 37; http://dx.doi.org/10.1074/jbc.M109446200; PMID: 11704675
- Görlach M, Burd CG, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res 1994; 211:400 - 7; http://dx.doi.org/10.1006/excr.1994.1104; PMID: 7908267
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005; 433:477 - 80; http://dx.doi.org/10.1038/nature03205; PMID: 15690031
- Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol 2008; 181:293 - 307; http://dx.doi.org/10.1083/jcb.200710215; PMID: 18426977
- Kumar GR, Glaunsinger BA. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol 2010; 30:4996 - 5008; http://dx.doi.org/10.1128/MCB.00600-10; PMID: 20823266
- Kühn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta 2004; 1678:67 - 84; PMID: 15157733