Abstract
Eukaryotic cells ensure error-free progress through the cell cycle by monitoring (1) the completion of cell cycle events, (2) damage to critical cellular components, or (3) structural changes such as the attachment of kinetochores to the mitotic spindle. In the presence of damage, or in the face of a reduced capacity to complete essential events, cells are capable of delaying the cell cycle so that damage can be repaired, or previous cell cycle phases can proceed to completion. Although such “checkpoints” have been extensively studied in many organisms—and much is understood with respect to the monitoring of DNA replication and DNA damage—little is known with regards to mechanisms that might monitor the completion of cytokinesis. In this review I summarize recent work from the fission yeast, Schizosaccharomyces pombe, describing the existence of regulatory modules that aid in ensuring the faithful and reliable execution of cytokinesis. Together, these modules promote the maintenance of a “cytokinesis-competent” state characterized by delayed progression into mitosis and the continuous repair and/or re-establishment of the acto-myosin ring. In this way, fission yeast cells are able to increase the likelihood of successful cell division prior to committing to a subsequent cell cycle. The recent demonstration of conservation between S. pombe components of these modules, and human proteins with defined roles in preventing cell division failure, suggest that the lessons learned in S. pombe may be applicable to other eukaryotes.
The Fission Yeast, Schizosaccharomyces pombe
The fission yeast, Schizosaccharomyces pombe, has been used extensively as a model system for the molecular-level understanding of a diverse array of biological processes (e.g., cell cycle control, RNA interference, cytoskeletal dynamics, DNA damage/repair).Citation1-Citation7 Interestingly, of the ~5000 genes in S. pombe, at least 172 share significant similarity to human genes whose mis-regulation leads to disease, and at least 23 share significant similarity to human genes with roles in the development of cancer.Citation6 Thus, S. pombe is an excellent model with which to understand the complex regulation of genetic networks and the consequences of their mis-regulation. Nowhere has this been more apparent than in the study of cytokinesis, where a multitude of recent studies have dramatically increased our knowledge of the mechanisms governing this fundamental process.Citation5,Citation8-Citation12 In many respects these studies have not only increased our knowledge regarding cytokinesis, but have also increased our general understanding of (1) how eukaryotic cells assemble and regulate complex genetic networks and (2) how these regulatory modules relate to higher order biological phenomenon.
While much is now known regarding the assembly and constriction of the actomyosin ring, our understanding of the mechanisms (if any) monitoring the completion of cytokinesis is lacking. In this review, I first present a brief summary of the regulatory modules required for the proper spatial and temporal regulation of cytokinesis in fission yeast. Next, I present an in-depth account of evidence that supports the existence of genetic networks with roles in promoting the reliable execution of cell division.
Spatial and Temporal Regulation of Cytokinesis in Fission Yeast
Cytokinesis comprises the period of the cell cycle in which newly segregated chromosomes are irreversibly separated into two independent daughter cells. While the absence of cytokinesis is tolerated under certain specialized circumstances—the development of Drosophila embryos, for example—it is normally essential for the proliferation and differentiation of actively growing cellular populations.Citation13,Citation14 In addition, recent work has also established that cytokinesis failure has dire consequences with respect to the maintenance of genomic integrity.Citation15-Citation17
Cytokinetic Actomyosin Ring Assembly
In fission yeast, just as in more developmentally complex eukaryotes, cytokinesis is achieved through the regulated assembly and subsequent constriction of a contractile actomyosin ring. The fission yeast actomyosin ring includes two type II myosin heavy chains, Myo2p and Myp2, together with their associated light chains, Cdc4p (essential light chain) and Rlc1p (regulatory light chain). In addition, the ring includes the IQGAP related protein, Rng2p, the PCH domain protein, Cdc15p, Cdc12p (formin), Cdc8p (tropomyosin), Cdc3p (profilin), as well as the actin filament cross linking proteins, Ain1p (α-actinin) and Fim1p (fimbrin). Mutations in essential actomyosin ring genes (e.g., cdc3, cdc4, cdc8, cdc12, rng2 and myo2) lead to the so-called “rng” phenotype; a phenotype characterized by the inability to assemble proper actomyosin rings and the subsequent appearance of elongated and branched cells with multiple nuclei and disorganized septal depositions.Citation8,Citation11,Citation12,Citation18
Contractile rings form from pre-cursor “interphase nodes” containing the analin-like protein Mid1p, the kinesin, Klp8 and the Rho family guanine nucleotide exchange factor, Gef2. Mid1p, which exits the nucleus and positions itself at the medial cortex prior to mitosis, acts as a “marker” for the future site of cell division (see below).Citation19 In addition to Mid1p, a group of regulatory kinases, Cdr1p, Cdr2p and Wee1p, are also present in interphase nodes. These kinases, together with the DYRK kinase, Pom1p, play a role in co-ordinating ring constriction with entry into mitosis.Citation8,Citation10,Citation19-Citation24
Pom1p localizes to the cell tips where it inhibits the Cdr1p and Cdr2p kinases and aids in restricting Mid1p to the medial region of the cell. As newly divided S. pombe cells grow in length—and the population of Pom1p molecules moves further from the medial region—this inhibition is relieved, allowing Cdr1p and Cdr2p to negatively regulate the Wee1p kinase through phosphorylation. In the presence of this negative regulation Wee1p is no longer able to inhibit the function of the cyclin dependent kinase, Cdc2p, thereby promoting the G2 to M transition. Thus, this elegant system allows for the proper spatial positioning of the ring, as well as for the co-ordination of ring assembly with entry into mitosis.Citation25-Citation28
Upon entry into mitosis the interphase nodes mature into “cytokinesis nodes” by the sequential recruitment of myosin II (along with its light chains), Rng2p, Cdc15p and Cdc12p. Once recruited, Cdc12p (formin), together with Cdc3p (profilin), promote actin filament polymerization. With the aid of tropomyosin, fimbrin and α-actinin, the actin filaments condense into an organized, bundled ring. Two non-exclusive models—the “leading cable” model and the “search, capture, pull and release” model—have been proposed to explain the condensation and constriction process.Citation8,Citation10-Citation12,Citation20-Citation24
The Septation Initiation Network
Once formed, the timing of ring constriction is controlled by an elaborate signaling pathway known as the septation initiation network (SIN). This network localizes to the spindle pole body (SPB; the centrosome equivalent in yeast) and triggers ring constriction.Citation9,Citation29,Citation30 In the absence of SIN signaling the actomyosin ring forms upon entry into mitosis, but then disassembles prematurely in late anaphase.Citation9,Citation18,Citation31-Citation34
The Spg1p GTPase acts as the “on-switch” of the SIN. Spg1p is negatively regulated by the two component GTPase activating protein (GAP), Cdc16p-Byr4p. Overexpression of Spg1p, or deletion of either Cdc16p or Byr4p, leads to multiple rounds of ring and septum formation without cell separation and intervening nuclear divisions.Citation9,Citation35-Citation39 When in its GTP bound form Spg1p promotes the recruitment of the Cdc7p protein kinase, which in-turn recruits the Sid1p-Cdc14p protein kinase complex to the SPB.
The scaffold proteins, Sid4p and Cdc11p also play a crucial role in SIN signaling as they function together in the recruitment of SIN components to the SPB. The most downstream component of the SIN is the Sid2p-Mob1p kinase complex whose enzymatic activity peaks during cytokinesis in a SIN dependent manner. This complex localizes to the SPB throughout the cell cycle, but also accumulates at the division site immediately prior to cytokinesis and is thought to transmit the division signal from the SPB to the actomyosin ring triggering its constriction.Citation9,Citation29,Citation30,Citation33,Citation40-Citation47 In addition to controlling ring constriction, more recent work has suggested that the SIN may also play a role in ring assembly through the recruitment of Cdc15p.Citation48
Regulatory Networks Governing the Linkage between Mitosis and Cytokinesis in S. pombe
In fission yeast, it is clear that the initiation of cytokinesis is linked to the completion of the preceding mitosis. This has been demonstrated by experiments involving the spindle checkpoint effector, Mad2p, which, when overexpressed, induces a metaphase arrest with increased levels of the cyclin dependent kinase, Cdc2p.Citation49 Under these conditions septation is normally prevented, however, in backgrounds where the activity of Cdc2p is reduced, cells are competent to form septa despite their failure to exit mitosis.Citation31
The mechanism by which this dependency is enforced involves the SIN components Cdc14p and Sid1p. While the overexpression of Mad2p has no effect on the localization of upstream SIN components, it does prevent the localization of Cdc14p and Sid1p to the SPB. Furthermore, when assayed directly, Cdc14p-Sid1p localization to the SPB occurs subsequent to Cdc2p kinase inactivation.Citation41 These data suggest that the localization of Sid1p-Cdc14p to the SPB, and thus activation of the SIN, is linked to mitotic exit through regulated inactivation of Cdc2p kinase activity.
Regulatory Networks Governing the Faithful Execution of Cytokinesis in S. pombe
While the mechanism described above ensures that cytokinesis only begins upon completion of the previous mitosis, it does not ensure that the completion of cytokinesis precedes entry into the subsequent mitosis. The remainder of this review will focus on the mechanisms present in fission yeast which maintain this dependency.
“Group I” vs. “Group II” Cytokinesis Mutants
The existence of a cytokinesis monitoring system was first suggested by experiments in which the nuclear cycle kinetics of temperature sensitive cytokinesis mutants were carefully analyzed.Citation50-Citation52 These experiments clearly revealed two classes of mutant behavior. While SIN mutants accumulate nuclei at a rate indistinguishable from wild-type cells at the restrictive temperature, rng mutants accumulate nuclei at a reduced rate. Furthermore, the delay seen in rng mutants is abolished in SIN mutant backgrounds. This indicates that rng mutants are being affected by a SIN dependent mechanism capable of delaying cell cycle progression. Cytokinesis mutants can thus be classified into two groups: Group I (comprising mutants that exhibit a delay in nuclear cycle progression upon cytokinetic perturbation), and Group II (comprising the SIN mutants, which proceed with the nuclear cycle unhindered upon failure in cytokinesis).Citation53
The Clp1p/Flp1p Phosphatase
The Cdc14p family of phosphatases is comprised of a highly conserved group of cell cycle regulators that function, at least in part, through dephosphorylating Cdc2p substrates.Citation54 Interestingly, the fission yeast Cdc14p family ortholog, Clp1p/Flp1p, has been strongly implicated as being a key regulator of the cytokinesis monitoring system. First, unlike rng single mutants (which are delayed in nuclear cycle progression), clp1Δ rng double mutants exhibit normal nuclear cycle kinetics upon shift to the restrictive temperature (thus placing the clp1 gene deletion in the type II mutant category).Citation50,Citation52 Second, clp1Δ mutants maintain a smaller cell size than wild type. This so called “semi-wee” phenotype is displayed by cells in which the transition from G2- to M-phase is accelerated and is a strong indication that Clp1p acts as a negative regulator of Cdc2p in promoting mitotic entry. Third, in contrast to clp1Δ and rng single mutants—which are able to grow and form colonies at semi-permissive temperatures—clp1Δ rng double mutants fail in cytokinesis and are inviable under these same growth conditions.Citation53 Lastly, upon entry into mitosis, a sub- population of Clp1p re-localizes from the nucleolus to the cytokinetic ring (as well as to the cytoplasm and the mitotic spindle).Citation50,Citation52 Taken together, these data point to Clp1p as being a likely candidate for maintaining the cell cycle delay observed upon perturbation of cell division structures, and show that clp1Δ cells are hyper-sensitive to a variety of mutations that disrupt the function of the actomyosin ring. Further evidence extending these observations—and showing that Clp1p confers a definitive survival advantage—has come from experiments utilizing low doses of the actin depolymerizing drug, latrunculin A (LatA).
LatA is a commonly used drug that acts by sequestering actin monomers.Citation55 At high concentrations (20–50 μM) this action prevents the re-assembly of monomers into actin filaments and results in the complete abrogation of F-actin structures within 20 min (and ultimately in cell death).Citation55,Citation56 However, when used at low concentrations (0.2–0.5 μM), the drug can be used as a tool to mildly perturb the ring and activate the cytokinesis monitoring system. For example, clp1Δ cells (synchronized in early G2 by centrifugal elutriation) enter the first mitosis and form actomyosin rings with similar kinetics to wild-type upon low dose LatA treatment. However, in contrast to similarly treated wild-type cells—which maintain the integrity of the ring for up to 120 min—clp1Δ mutants exhibit ring fragmentation/disassembly within only 30 min. Furthermore, whereas wild-type cells are able to delay mitotic entry (of the second mitosis) until the actomyosin ring slowly constricts, clp1Δ cells proceed with the second mitosis (becoming tetra-nucleate) with kinetics indistinguishable from mock treated controls.Citation53 Moreover, while clp1Δ cells are inviable on solid media containing low dose LatA, wild-type cells are capable of growth and colony formation.Citation53 Thus, consistent with earlier genetic analysis, these data strongly indicate that Clp1p mediates a survival advantage upon LatA treatment through both delaying cell cycle progression and by stabilizing the actomyosin ring.
A Dual Role for Clp1p in Preventing Cytokinesis Failure
An intriguing question that arises when considering the cytokinesis monitoring system concerns the relationship between Clp1p mediated cell cycle arrest and actomyosin ring stability. Clp1p plays a definitive role in regulating the transition from interphase to mitosis. Thus, is increased stability of the actomyosin ring simply the indirect consequence of cell cycle delay, or does Clp1p play a direct role in stabilizing the ring?
To answer this question temperature-sensitive mutations in the mitotic inducer cdc25 can be used to provide a Clp1p independent cell cycle block upon low dose LatA treatment. Using this strategy it is thus possible to assay actomyosin ring stability under conditions where Clp1p activity is absent, but an interphase arrest is still in effect. Intriguingly, actomyosin rings in cdc25-22 clp1Δ cells, unlike cdc25-22 controls, do indeed fragment in the presence of low doses of LatA.Citation53 Thus, Clp1p functions not only to delay cell cycle progression, but also to maintain actomyosin ring integrity. These two functions complement one another, providing the cell with a lengthened duration in which to properly constrict the actomyosin ring and successfully complete cytokinesis. It is of particular interest to note the similarity between this bimodal mechanism and other bimodal responses in other characterized checkpoints. For example, in budding yeast, the checkpoint kinase Mec1p is required not only to stabilize slowed or stalled replication forks, but also to delay progress through S phase.Citation57
Clp1p Increases the Duration of SIN Signaling
One of the most intriguing aspects of SIN mutants is their ability to form, but not constrict actomyosin rings. Upon shift to the restrictive temperature SIN mutants form rings which are similar to those observed in wild-type cells, however, unlike wild-type cells, these rings disassemble in late anaphase resulting in cytokinesis failure.Citation58,Citation59 This phenotype is remarkably similar to the phenotype displayed by clp1Δ cells upon treatment with low doses of LatA. Under these conditions clp1Δ cells are also competent to form actomyosin rings, but these rings disassemble in a similar fashion to SIN mutants prior to constriction.Citation53
To help shed light on the relationship between Clp1p and the SIN, the localization of SIN components (Cdc7p and Sid1p) were examined upon low dose LatA treatment in clp1Δ and wild-type backgrounds.Citation53 Cdc7p and Sid1p localize to a single SPB during late mitosis, and remain there until the completion of cytokinesis. Thus, the localization of these components to the SPB serves as a marker of active SIN signaling.Citation60
Interestingly, these studies echoed results obtained when examining actomyosin ring stability. Cdc7p and Sid1p localize to the SPB with similar kinetics in synchronized, LatA treated wild-type and clp1Δ cells. However, while wild-type cells are able to maintain Cdc7p and Sid1p to the SPB for prolonged periods of time (up to 240 min), clp1Δ cells display a dramatic drop in the percentage of cells with these components at the SPB.Citation53 In addition, ring fragmentation in low dose LatA treated clp1Δ cells can be rescued by mutations resulting in hyper-activation of the SIN.Citation53 Thus, taking all data together, these results strongly suggest that Clp1p functions to extend the duration of SIN signaling upon cytokinetic perturbations, thereby providing an improved opportunity to properly constrict the ring.
Interestingly, the Clp1p dependent maintenance of SIN activity complements work examining the DNA damage checkpoint in fission yeast. In this work the authors demonstrate that checkpoint arrest in the presence of DNA damage must be actively maintained through the function of the Chk1p protein kinase.Citation61 This parallels the function of Clp1p in the sense that Clp1p is not required to activate the SIN (clp1Δ mutants are fully capable of constricting rings and forming septa under normal growth conditions), but is important in extending the duration of SIN signaling (as needed) in the presence of damage to the cell division machinery.
Clp1p and the SIN function in a Positive Feedback Loop
Like members of the SIN, Clp1p displays cell cycle regulated changes in its sub-cellular localization during normal logarithmic growth. In interphase, Clp1p is present in the nucleolus and SPB. However, upon mitotic entry, Clp1p re-localizes to the cytoplasm/mitotic spindle/actomyosin ring and remains at these locations until the completion of cytokinesis.Citation50,Citation52
Intriguingly, upon LatA treatment Clp1p is retained in the cytoplasm for prolonged periods of time. This retention is mediated through the action of the SIN kinase, Sid2p, and the 14-3-3 protein, Rad24p. Upon SIN activation, Sid2p phosphorylates Clp1p creating a binding site for the Rad24p, protein. The interaction between Rad24p and Clp1p thus permits retention of Clp1p in the cytoplasm (). As expected, rad24Δ cells are unable to maintain prolonged SIN signaling upon LatA treatment leading to inviability and a terminal phenotype indistinguishable from clp1Δ cells.Citation62,Citation63
Figure 1. Regulatory dynamics during cytokinesis in S. pombe. (A) During interphase, Mbx1p, a component of the pombe cell cycle box binding factor (PBF), is bound and dephosphorylated by Clp1p. In this form Mbx1p aids in repression of the transcription of a set of genes (including mid1, sid2, and cdc15) with roles in cytokinesis. (B) During normal cytokinesis—or upon extension of the cytokinetic (or “C”) phase of the cell cycle upon perturbation of the cell division machinery—Plo1p phosphorylates Mbx1p, relieving its inhibitory effect on the transcription of cytokinesis genes such as sid2. Sid2p phosphorylates Clp1p, creating a binding site for the 14-3-3 protein, Rad24p. Rad24p binding to Clp1p promotes the cytoplasmic retention of Clp1p, preventing it from de-phosphorylating Mbx1p.
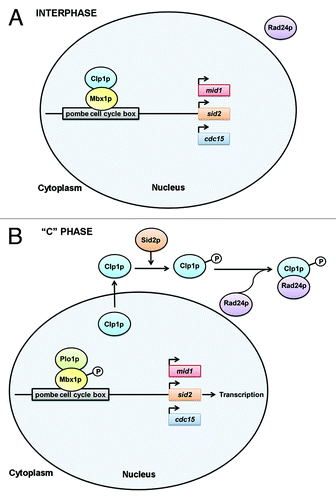
Thus, upon cell division stress, SIN signaling is required for the prolonged maintenance of Clp1p in the cytoplasm, and Clp1p is required for the prolonged maintenance of SIN signaling. These data therefore strongly support a model in which Clp1p and the SIN act in a positive feedback loop (cytoplasmic Clp1p promotes SIN activity and active SIN promotes retention of Clp1p in the cytoplasm). In this way the cytokinetic phase of the cell cycle may be extended and cell cycle progress delayed. Together these mechanisms promote the successful constriction of the actomyosin ring prior to the initiation of mitosis.
Regulation of Gene Expression by Clp1p
Interestingly, Clp1p may also aid in maintaining a “cytokinesis competent” state through affecting cell cycle dependent gene-expression. In S. pombe, a trans-acting transcription factor complex (pombe cell cycle box binding factor or PBF) controls a wave of transcription at the M-G1 interval. Not surprisingly, the genes in this wave encode proteins (including Sid2p and Mid1p) with roles in cytokinesis and cell division. Remarkably, one component of this complex, Mbx1p, is a substrate of Clp1p.Citation64 When present in the nucleus, Clp1p, binds to and de-phosphorylates Mbx1p, thereby aiding in the repression of transcription of the M-G1 wave. In contrast, when Clp1p is present in the cytoplasm, Mbx1p is free to be phosphorylated (by the polo family kinase, Plo1p), thus stimulating transcription of the M-G1 wave.Citation64,Citation65 Thus, this transcriptional mechanism provides another avenue by which the cytoplasmic retention of Clp1p promotes a “cytokinesis competent” state conducive to the successful completion of cell division ().
Genome Wide Screens Identify Novel Regulatory Modules with Roles in Preventing Cytokinesis Failure
As described above, the role of regulators such as Clp1p and Rad24p in monitoring cytokinesis can be revealed experimentally through the treatment of gene deletion mutants with low doses of LatA. Thus, comprehensive and unbiased genome-wide screens – based on hyper-sensitivity to LatA—can be used to identify novel regulatory components of genetic networks with roles in promoting successful cell division. Just such a screen identified the lsk1 (latrunculin sensitive kinase knockout) gene. Similar to clp1Δ cells, lsk1Δ mutants appear normal under typical growth conditions, but display severe cytokinesis defects upon perturbation of the cell division machinery. Furthermore—as shown by genetic analysis with various hypo- and hyper-active SIN mutants—Lsk1p acts as a positive regulator of the SIN.Citation66
Somewhat surprisingly—given its role in regulating cytokinesis—the lsk1 gene encodes a kinase that specifically phosphorylates the Ser-2 residues present in the heptad repeats (Y1S2P3T4S5P6S7) of the carboxy terminal domain (CTD) of the largest subunit of RNA polymerase II, Rpb1p.Citation67 Lsk1p displays significant sequence similarity to human Cdk9p, which, together with cyclin T, forms the p-TEFb complex that targets Ser-2 residues of the RNA pol II CTD and modulates pre-mRNA processing.Citation67,Citation68 Importantly, S. pombe cells bearing rpb1 alleles (rpb1-12XS2ACTD) encoding 12 copies of a mutant heptad in which alanine is substituted for the second serine (in order to mimic the non-phosphorylated state) display cytokinesis phenotypes indistinguishable from lsk1Δ strains.Citation69 These data are consistent with a model in which the Ser-2 residues of the CTD are indeed the biologically relevant targets of Lsk1p with respect to cytokinesis. Furthermore, these data suggest that upstream kinases might act through the CTD to selectively control the transcription of certain gene subsets.
Interestingly, while gene expression profiling of mutants impaired in Ser-2 phosphorylation display little change with respect to the transcription of most genes, a small sub-set of genes affecting cytokinesis—including the actin interacting protein, Aip1p, the Clp1p interacting protein, Nsk1p, and the SPB protein, Cut12p—were found to be differentially regulated.Citation70 The identification of these genes (particularly Nsk1p, a known interactor of the critical Clp1p phosphatase), together with genetic data showing that Lsk1p is a positive regulator of SIN signaling, suggests a model in which Lsk1p dependent modulation of transcription may also contribute to maintaining a “cytokinesis competent” state.
The final group of regulators to be discussed define components of a putative histone de-acetylase (HDAC) complex. Identified through a comprehensive genome wide screen of gene deletion mutants, Set3p, Snt1p, and Hif2p, are required to prevent cytokinesis failure in response to LatA treatment.Citation71 Moreover, consistent with a role in responding to LatA induced damage to the cell division machinery, the protein levels of all three regulators increase 2–3 fold upon challenge with the drug.Citation71
While global gene expression profiling of set3Δ cells revealed that cytokinesis genes were not affected, the analysis did show that Set3p was required to regulate genes with roles in the cellular response to stress. In contrast to wild-type cells, which respond to LatA treatment with strong induction/repression of the core environmental stress response genes (~60% of the CESR genes differentially regulated), set3Δ mutants are unable to properly modulate the CESR genes (only 1% of the CESR genes differentially regulated).Citation71,Citation72 Thus, cytokinetic failure in set3Δ cells may be a manifestation of the mutant's inability to properly adapt to the presence of LatA leading to direct and/or indirect effects on the function of the cytokinetic machinery.
When considering this last group of regulators it is particularly intriguing to note that orthologs of Hif2p, Set3p, and Snt1p exist in humans (TBL1X, MLL5, and NCOR2, respectively). Furthermore—as might be expected based on the selection criteria used in the fission yeast screen—these human orthologs have themselves been shown to play a role in cytokinesis in human cells.Citation73 In this study the researchers discovered that the knockdown of either MLL5, TBL1X, or NCOR2 resulted in defects in furrow ingression, subsequent cytokinesis failure, and finally the generation of bi-nucleate (i.e., tetraploid) intermediates with twice the normal number of centrosomes.
Final Thoughts
The importance of understanding the mechanisms utilized by eukaryotes to ensure the dependable execution of cytokinesis was first articulated by Theodor Boveri almost 100 y ago.Citation74 In his classic work “Concerning the origin of malignant tumours” Boveri hypothesized that tetraploid intermediates—derived from either cytokinetic failure or cell fusion—might undergo chaotic multipolar mitoses leading to numerical and/or structural chromosomal defects.
Recent evidence provides experimental support for Boveri’s ideas. For instance, tetraploid mouse mammary epithelial cells generated by the inhibition of cytokinesis display increased rates of aneuploidy and (when transplanted into nude mice) give rise to malignant tumors at greater rates than controls.Citation15-Citation17,Citation75 These results suggest that mechanisms promoting the dependable execution of cytokinesis may be important in maintaining genomic integrity and in preventing carcinogenesis. Thus, in the broadest sense, an understanding of these pathways may provide a better understanding of one “route” by which eukaryotic cells become tumorigenic in multicellular organisms (for reviews on this subject and a detailed discussion of further experimental evidence please see refs. Citation15–Citation17).
This last thought raises the question of whether these regulatory modules—which all seem to promote and/or aid in establishing a cellular state conducive to cytokinesis—are specific to S. pombe, or whether they may also be of relevance to cytokinesis in more developmentally complex eukaryotes? While highly speculative, some work in mammalian cells suggests that a monitoring system may very well exist in higher eukaryotes. For example cells deficient for phosphotidylethanolamine are unable to complete cytokinesis, and arrest with a stable actomyosin ring and interphase nuclei.Citation76 Furthermore, mammalian cells treated with a myosin II ATPase inhibitor also arrest with two nuclei and a stable actomyosin ring.Citation77 In any event, future work will undoubtedly continue to expand our knowledge regarding these regulatory systems and provide further insight into their molecular nature and their relationship to genomic stability.
| Abbreviations: | ||
| SIN | = | septation initiation network |
| LatA | = | latrunculin A |
| SPB | = | spindle pole body |
| PCB | = | pombe cell cycle box |
| PBF | = | pombe cell cycle box binding factor |
| CTD | = | carboxy-terminal domain |
| CESR | = | core environmental stress response |
| GAP | = | GTPase activating protein |
| PCH | = | pombe cdc15 homology |
| IQGAP | = | IQ motif and GTPase activating domain |
| DYRK | = | dual-specificity tyrosine-regulated kinase |
Acknowledgments
We would like to thank members of the UWO Biology and Biochemistry departments for helpful discussions and/or critical reading of the manuscript.
References
- Chang F, Martin SG. Shaping fission yeast with microtubules. Cold Spring Harb Perspect Biol 2009; 1:a001347; http://dx.doi.org/10.1101/cshperspect.a001347; PMID: 20066076
- Dang Y, Yang Q, Xue Z, Liu Y. RNA interference in fungi: pathways, functions, and applications. Eukaryot Cell 2011; 10:1148 - 55; http://dx.doi.org/10.1128/EC.05109-11; PMID: 21724934
- Kovar DR, Sirotkin V, Lord M. Three’s company: the fission yeast actin cytoskeleton. Trends Cell Biol 2011; 21:177 - 87; http://dx.doi.org/10.1016/j.tcb.2010.11.001; PMID: 21145239
- Kuntz K, O’Connell MJ. The G(2) DNA damage checkpoint: could this ancient regulator be the Achilles heel of cancer?. Cancer Biol Ther 2009; 8:1433 - 9; http://dx.doi.org/10.4161/cbt.8.15.9081; PMID: 19574738
- McInerny CJ. Cell cycle regulated gene expression in yeasts. Adv Genet 2011; 73:51 - 85; http://dx.doi.org/10.1016/B978-0-12-380860-8.00002-1; PMID: 21310294
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002; 415:871 - 80; http://dx.doi.org/10.1038/nature724; PMID: 11859360
- Yanagida M. The model unicellular eukaryote, Schizosaccharomyces pombe. Genome Biol 2002; 3:T2003; http://dx.doi.org/10.1186/gb-2002-3-3-comment2003; PMID: 11897018
- Bathe M, Chang F. Cytokinesis and the contractile ring in fission yeast: towards a systems-level understanding. Trends Microbiol 2010; 18:38 - 45; http://dx.doi.org/10.1016/j.tim.2009.10.002; PMID: 19959363
- Goyal A, Takaine M, Simanis V, Nakano K. Dividing the spoils of growth and the cell cycle: The fission yeast as a model for the study of cytokinesis. Cytoskeleton (Hoboken) 2011; 68:69 - 88; http://dx.doi.org/10.1002/cm.20500; PMID: 21246752
- Laporte D, Zhao R, Wu JQ. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin Cell Dev Biol 2010; 21:892 - 8; http://dx.doi.org/10.1016/j.semcdb.2010.08.004; PMID: 20708088
- Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol 2010; 22:50 - 6; http://dx.doi.org/10.1016/j.ceb.2009.11.010; PMID: 20031383
- Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol 2010; 11:149 - 55; http://dx.doi.org/10.1038/nrm2834; PMID: 20094054
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol 2004; 14:R806 - 18; http://dx.doi.org/10.1016/j.cub.2004.09.022; PMID: 15380095
- Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev 2002; 66:155 - 78; http://dx.doi.org/10.1128/MMBR.66.2.155-178.2002; PMID: 12040122
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev 2007; 17:157 - 62; http://dx.doi.org/10.1016/j.gde.2007.02.011; PMID: 17324569
- Pellman D. Cell biology: aneuploidy and cancer. Nature 2007; 446:38 - 9; http://dx.doi.org/10.1038/446038a; PMID: 17330036
- Sagona AP, Stenmark H. Cytokinesis and cancer. FEBS Lett 2010; 584:2652 - 61; http://dx.doi.org/10.1016/j.febslet.2010.03.044; PMID: 20371245
- Le Goff X, Utzig S, Simanis V. Controlling septation in fission yeast: finding the middle, and timing it right. Curr Genet 1999; 35:571 - 84; http://dx.doi.org/10.1007/s002940050455; PMID: 10467001
- Almonacid M, Paoletti A. Mechanisms controlling division-plane positioning. Semin Cell Dev Biol 2010; 21:874 - 80; http://dx.doi.org/10.1016/j.semcdb.2010.08.006; PMID: 20708089
- Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 2008; 319:97 - 100; http://dx.doi.org/10.1126/science.1151086; PMID: 18079366
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science 2005; 310:310 - 4; http://dx.doi.org/10.1126/science.1113230; PMID: 16224022
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, et al. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol 2006; 174:391 - 402; http://dx.doi.org/10.1083/jcb.200602032; PMID: 16864655
- Yonetani A, Chang F. Regulation of cytokinesis by the formin cdc12p. Curr Biol 2010; 20:561 - 6; http://dx.doi.org/10.1016/j.cub.2010.01.061; PMID: 20226666
- Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell 2008; 19:2208 - 19; http://dx.doi.org/10.1091/mbc.E07-07-0731; PMID: 18305104
- Huang Y, Chew TG, Ge W, Balasubramanian MK. Polarity determinants Tea1p, Tea4p, and Pom1p inhibit division-septum assembly at cell ends in fission yeast. Dev Cell 2007; 12:987 - 96; http://dx.doi.org/10.1016/j.devcel.2007.03.015; PMID: 17543869
- Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature 2009; 459:852 - 6; http://dx.doi.org/10.1038/nature08054; PMID: 19474792
- Padte NN, Martin SG, Howard M, Chang F. The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr Biol 2006; 16:2480 - 7; http://dx.doi.org/10.1016/j.cub.2006.11.024; PMID: 17140794
- Saunders TE, Pan KZ, Angel A, Guan Y, Shah JV, Howard M, et al. Noise Reduction in the Intracellular Pom1p Gradient by a Dynamic Clustering Mechanism. Dev Cell 2012; http://dx.doi.org/10.1016/j.devcel.2012.01.001; PMID: 22342545
- Krapp A, Schmidt S, Cano E, Simanis VS. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr Biol 2001; 11:1559 - 68; http://dx.doi.org/10.1016/S0960-9822(01)00478-X; PMID: 11676915
- Morrell JL, Tomlin GC, Rajagopalan S, Venkatram S, Feoktistova AS, Tasto JJ, et al. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr Biol 2004; 14:579 - 84; http://dx.doi.org/10.1016/j.cub.2004.03.036; PMID: 15062098
- Fankhauser C, Marks J, Reymond A, Simanis V. The S. pombe cdc16 gene is required both for maintenance of p34cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis?. EMBO J 1993; 12:2697 - 704; PMID: 8334988
- Fankhauser C, Simanis V. The Schizosaccharomyces pombe cdc14 gene is required for septum formation and can also inhibit nuclear division. Mol Biol Cell 1993; 4:531 - 9; PMID: 8334307
- Fankhauser C, Simanis V. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J 1994; 13:3011 - 9; PMID: 8039497
- Simanis V. The control of septum formation and cytokinesis in fission yeast. Semin Cell Biol 1995; 6:79 - 87; http://dx.doi.org/10.1016/1043-4682(95)90004-7; PMID: 7548846
- Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev 1997; 11:1519 - 34; http://dx.doi.org/10.1101/gad.11.12.1519; PMID: 9203579
- Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol 1998; 8:947 - 54; http://dx.doi.org/10.1016/S0960-9822(98)70394-X; PMID: 9742395
- Cerutti L, Simanis V. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J Cell Sci 1999; 112:2313 - 21; PMID: 10381387
- Furge KA, Cheng QC, Jwa M, Shin S, Song K, Albright CF. Regions of Byr4, a regulator of septation in fission yeast, that bind Spg1 or Cdc16 and form a two-component GTPase-activating protein with Cdc16. J Biol Chem 1999; 274:11339 - 43; http://dx.doi.org/10.1074/jbc.274.16.11339; PMID: 10196225
- Krapp A, Collin P, Cano Del Rosario E, Simanis V. Homoeostasis between the GTPase Spg1p and its GAP in the regulation of cytokinesis in S. pombe. J Cell Sci 2008; 121:601 - 8; http://dx.doi.org/10.1242/jcs.022772; PMID: 18252797
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol 1999; 146:777 - 90; http://dx.doi.org/10.1083/jcb.146.4.777; PMID: 10459013
- Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J 2000; 19:1803 - 15; http://dx.doi.org/10.1093/emboj/19.8.1803; PMID: 10775265
- Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol 2000; 10:619 - 22; http://dx.doi.org/10.1016/S0960-9822(00)00492-9; PMID: 10837231
- Guertin DA, McCollum D. Interaction between the noncatalytic region of Sid1p kinase and Cdc14p is required for full catalytic activity and localization of Sid1p. J Biol Chem 2001; 276:28185 - 9; http://dx.doi.org/10.1074/jbc.M103802200; PMID: 11384993
- Hou MC, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol Cell Biol 2004; 24:3262 - 76; http://dx.doi.org/10.1128/MCB.24.8.3262-3276.2004; PMID: 15060149
- Krapp A, Cano E, Simanis V. Analysis of the S. pombe signalling scaffold protein Cdc11p reveals an essential role for the N-terminal domain in SIN signalling. FEBS Lett 2004; 565:176 - 80; http://dx.doi.org/10.1016/j.febslet.2004.03.098; PMID: 15135075
- Singh NS, Shao N, McLean JR, Sevugan M, Ren L, Chew TG, et al. SIN-inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates SIN asymmetry in fission yeast. Curr Biol 2011; 21:1968 - 78; http://dx.doi.org/10.1016/j.cub.2011.10.051; PMID: 22119525
- Johnson AE, Gould KL. Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase. EMBO J 2011; 30:341 - 54; http://dx.doi.org/10.1038/emboj.2010.317; PMID: 21131906
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev 2008; 22:3205 - 16; http://dx.doi.org/10.1101/gad.1697208; PMID: 19056897
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci U S A 1997; 94:7965 - 70; http://dx.doi.org/10.1073/pnas.94.15.7965; PMID: 9223296
- Cueille N, Salimova E, Esteban V, Blanco M, Moreno S, Bueno A, et al. Flp1, a fission yeast orthologue of the s. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J Cell Sci 2001; 114:2649 - 64; PMID: 11683392
- Liu J, Wang H, Balasubramanian MK. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J Cell Sci 2000; 113:1223 - 30; PMID: 10704373
- Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol 2001; 11:931 - 40; http://dx.doi.org/10.1016/S0960-9822(01)00268-8; PMID: 11448769
- Mishra M, Karagiannis J, Trautmann S, Wang H, McCollum D, Balasubramanian MK. The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J Cell Sci 2004; 117:3897 - 910; http://dx.doi.org/10.1242/jcs.01244; PMID: 15265986
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 1998; 2:709 - 18; http://dx.doi.org/10.1016/S1097-2765(00)80286-5; PMID: 9885559
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol 1997; 137:399 - 416; http://dx.doi.org/10.1083/jcb.137.2.399; PMID: 9128251
- Rupes I, Webb BA, Mak A, Young PG. G2/M arrest caused by actin disruption is a manifestation of the cell size checkpoint in fission yeast. Mol Biol Cell 2001; 12:3892 - 903; PMID: 11739788
- Pasero P, Shimada K, Duncker BP. Multiple roles of replication forks in S phase checkpoints: sensors, effectors and targets. Cell Cycle 2003; 2:568 - 72; http://dx.doi.org/10.4161/cc.2.6.577; PMID: 14512770
- Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, et al. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics 1998; 149:1265 - 75; PMID: 9649519
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev 1997; 11:2939 - 51; http://dx.doi.org/10.1101/gad.11.22.2939; PMID: 9367977
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol 2001; 11:89 - 95; http://dx.doi.org/10.1016/S0962-8924(00)01901-2; PMID: 11166217
- Latif C, den Elzen NR, O’Connell MJ. DNA damage checkpoint maintenance through sustained Chk1 activity. J Cell Sci 2004; 117:3489 - 98; http://dx.doi.org/10.1242/jcs.01204; PMID: 15213253
- Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM, Gould KL, et al. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol 2008; 18:1594 - 9; http://dx.doi.org/10.1016/j.cub.2008.08.067; PMID: 18951025
- Mishra M, Karagiannis J, Sevugan M, Singh P, Balasubramanian MK. The 14-3-3 protein rad24p modulates function of the cdc14p family phosphatase clp1p/flp1p in fission yeast. Curr Biol 2005; 15:1376 - 83; http://dx.doi.org/10.1016/j.cub.2005.06.070; PMID: 16085489
- Papadopoulou K, Chen JS, Mead E, Feoktistova A, Petit C, Agarwal M, et al. Regulation of cell cycle-specific gene expression in fission yeast by the Cdc14p-like phosphatase Clp1p. J Cell Sci 2010; 123:4374 - 81; http://dx.doi.org/10.1242/jcs.073056; PMID: 21098641
- McInerny CJ. Clp1p and Mid1p form links between cell cycle progression and gene expression at cytokinesis in fission yeast. Cell Cycle 2011; 10:1184 - 5; http://dx.doi.org/10.4161/cc.10.8.15346; PMID: 21512312
- Karagiannis J, Bimbó A, Rajagopalan S, Liu J, Balasubramanian MK. The nuclear kinase Lsk1p positively regulates the septation initiation network and promotes the successful completion of cytokinesis in response to perturbation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell 2005; 16:358 - 71; http://dx.doi.org/10.1091/mbc.E04-06-0502; PMID: 15537703
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 2006; 20:2922 - 36; http://dx.doi.org/10.1101/gad.1477006; PMID: 17079683
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 2006; 23:297 - 305; http://dx.doi.org/10.1016/j.molcel.2006.06.014; PMID: 16885020
- Karagiannis J, Balasubramanian MK. A cyclin-dependent kinase that promotes cytokinesis through modulating phosphorylation of the carboxy terminal domain of the RNA Pol II Rpb1p sub-unit. PLoS One 2007; 2:e433; http://dx.doi.org/10.1371/journal.pone.0000433; PMID: 17502918
- Saberianfar R, Cunningham-Dunlop S, Karagiannis J. Global gene expression analysis of fission yeast mutants impaired in Ser-2 phosphorylation of the RNA pol II carboxy terminal domain. PLoS One 2011; 6:e24694; http://dx.doi.org/10.1371/journal.pone.0024694; PMID: 21931816
- Rentas S, Saberianfar R, Grewal C, Kanippayoor R, Mishra M, McCollum D, et al. The SET Domain Protein, Set3p, Promotes the Reliable Execution of Cytokinesis in Schizosaccharomyces pombe. PLoS One 2012; 7:e31224; http://dx.doi.org/10.1371/journal.pone.0031224; PMID: 22347452
- Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, et al. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell 2003; 14:214 - 29; http://dx.doi.org/10.1091/mbc.E02-08-0499; PMID: 12529438
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol 2007; 9:1401 - 12; http://dx.doi.org/10.1038/ncb1659; PMID: 17994010
- Harris H. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. Preface. J Cell Sci 2008; 121:Suppl 1 v - vi; http://dx.doi.org/10.1242/jcs.025759; PMID: 18089651
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005; 437:1043 - 7; http://dx.doi.org/10.1038/nature04217; PMID: 16222300
- Emoto K, Umeda M. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J Cell Biol 2000; 149:1215 - 24; http://dx.doi.org/10.1083/jcb.149.6.1215; PMID: 10851019
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 2003; 299:1743 - 7; http://dx.doi.org/10.1126/science.1081412; PMID: 12637748