Abstract
Phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) is a key signaling molecule in chemotaxis, a directed cell migration toward chemoattractants. PtdIns(3,4,5)P3 is transiently generated by chemotactic stimulation and activates reorganization of the actin cytoskeleton at the leading edge of migrating cells. In a recent study, we demonstrated that PtdIns(3,4,5)P3 directly binds to three members of the actin-based motor protein myosin I (myosin ID, IE and IF) in Dictyostelium discoideum and recruits these proteins to the plasma membrane of the leading edge. The PtdIns(3,4,5)P3-regulated membrane recruitment of myosin I induced chemoattractant-stimulated actin polymerization and was therefore required for chemotaxis. Similarly, human myosin IF was translocated to the plasma membrane through interactions with PtdIns(3,4,5)P3 upon chemotactic stimulation in a neutrophil cell line. Interestingly, we also found that the three PtdIns(3,4,5)P3-binding myosin I proteins function in phagocytosis, which involves both PtdIns(3,4,5)P3 signaling and actin cytoskeleton remodeling. Our findings provide an evolutionarily conserved mechanism by which class I myosin transmits PtdIns(3,4,5)P3 signals to the actin cytoskeleton.
Chemotaxis plays important roles in a variety of biological processes, such as embryonic development, axon guidance, wound healing and immune responses. In addition to normal physiology, chemotactic migration is also linked to many pathological conditions. For instance, chemotaxis of tumor cells leads to cancer metastasis, while unwanted immune cell chemotaxis causes chronic inflammatory diseases.Citation1-Citation5 In chemotaxis, chemoattractants such as growth factors, insulin and chemokines bind to their receptors on the cell surface. The majority of chemoattractant receptors are either seven-transmembrane G-protein-coupled receptors or receptor tyrosine kinase receptors.Citation6-Citation8 Among downstream mechanisms of these receptors, the lipid molecule PtdIns(3,4,5)P3 plays critical roles in intracellular chemotactic signaling. PtdIns(3,4,5)P3 is produced in the plasma membrane by PI3-kinase (PI3K) and is turned over by the PI3-phosphatase [phosphatase and tensin homolog (PTEN)] or PI5-phosphatase (SHIP1).Citation9-Citation18 PI3Ks are activated upon chemotactic stimulation and are transiently associated with the plasma membrane at the leading edge of migrating cells. PI3Ks convert phosphatidylinositol (4,5)-bisphosphate (PtdIns(4,5)P2) to PtdIns(3,4,5)P3. Conversely, PTEN and SHIP1 degrade PtdIns(3,4,5)P3. Therefore, PI3K and PTEN/SHIP1 provide regulation of PtdIns(3,4,5)P3 production, with synthesis occurring at the leading edge and degradation at the rear.
To identify novel components that mediate PtdIns(3,4,5)P3 signaling in chemotaxis, we used a proteomic approach involving affinity purification of PtdIns(3,4,5)P3-binding proteins from Dictyostelium cytosol and their identification using mass spectrometry.Citation19 Our experiments identified five PH domain-containing proteins, including two previously characterized proteins, PhdA and PKB, and three novel proteins that we named PhdB, PhdG, and PhdI. We have shown that PhdB, PhdG and PhdI bind specifically to PtdIns(3,4,5)P3 through PH domains in vitro and in vivo, and that these proteins are functionally important for chemotaxis. In addition to PH domain-containing proteins, we identified three class I myosin proteins, including myosin ID, IE and IF.
In our recent study, we showed that myosin ID, IE and IF are required for chemotaxis using a gene knockout approach.Citation20 Cells lacking these class I myosins were defective in chemoattractant-stimulated actin polymerization. Myosin I is a monomeric, actin-based motor protein with ATPase activity and has been shown to function in membrane-cytoskeletal interactions, including vesicle transport along actin filaments and regulation of plasma membrane tension.Citation21-Citation23 Myosin I molecules also have a tail homology (TH) domain that contains a putative PH domain phosphatidylinositol-binding motif. Previous studies have shown that the TH domain preferentially binds to acidic phospholipids such as phosphatidylserine and PtdIns(4,5)P2. These phospholipids are relatively abundant in biological membranes and may not change their levels in response to intracellular signaling. In contrast, PtdIns(3,4,5)P3 levels are highly regulated and function as signaling mechanisms. Our finding that myosin ID, IE and IF interact with PtdIns(3,4,5)P3 suggests that these myosin molecules are regulated by PtdIns(3,4,5)P3.
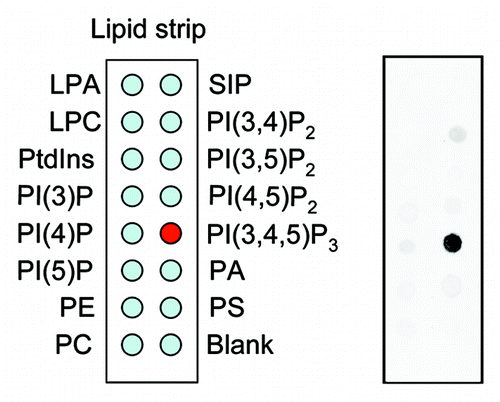
We demonstrated that myosin ID, IE and IF specifically bind to PtdIns(3,4,5)P3 in lipid dot-blot and liposome binding assays ().Citation20 For these assay, we expressed myosin I as green fluorescent protein (GFP) fusion proteins in Dictyostelium cells. The total cell lysate was incubated with nitrocellulose membranes or liposomes carrying different lipids, and interactions of myosin I-GFP with phospholipids were detected using anti-GFP antibodies. To determine whetehr these myosin I proteins directly bind to PtdIns(3,4,5)P3, we immunopurified myosin IE-GFP from Dictyostelium cells and incubated it with fluorescently labeled PtdIns(3,4,5)P3. In this in vitro binding assay, beads carrying myosin IE-GFP specifically associated with PtdIns(3,4,5)P3. The PtdIns(3,4,5)P3-myosin I interactions are mediated by TH1 domain as mutations in this domain abolished the lipid-protein interaction. Furthermore, we showed that human myosin IF, which has been suggested to bind to PtdIns(3,4,5)P3 in a previous proteomic study,Citation24 also binds to PtdIns(3,4,5)P3 in a lipid dot-blot assay, and that mutations in the TH domain of human myosin IF blocked its interaction with PtdIns(3,4,5)P3. These data indicate that the ability of TH domain-containing myosin I to bind to PtdIns(3,4,5)P3 is evolutionarily conserved.
Figure 1. Lipid dot blot assay. Whole cell lysates were prepared from Dictyostelium cells expressing myosin IE fused to green fluorescent protein and incubated with nitrocellulose membranes carrying different phosphatidylinositols (Echelon). Lipid–protein interactions were detected with anti-GFP antibodies.
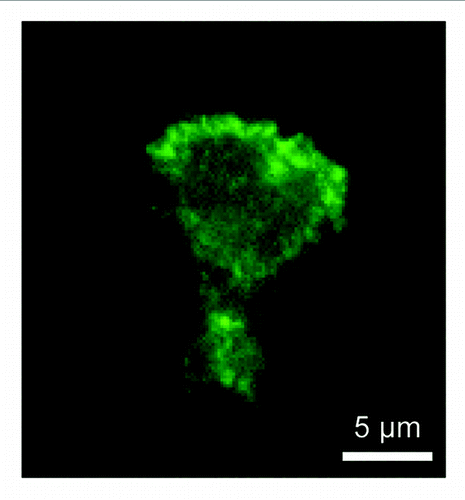
In chemotaxing cells, PtdIns(3,4,5)P3-binding myosin I is located at the leading edge.Citation20 This localization is mediated by interactions with PtdIns(3,4,5)P3 since mutations that block PtdIns(3,4,5)P3 interactions inhibited myosin I's localization at the leading edge and function in chemoattractant-stimulated actin polymerization, resulting in chemotaxis defects. Similarly, human myosin IF fused to yellow fluorescent protein (YFP-myosin IF) was recruited to the plasma membrane in COS-7 cells upon stimulation with epidermal growth factor, which increases PtdIns(3,4,5)P3 levels in the plasma membrane. When expressed in the human neutrophil cell line HL-60, YFP-myosin IF was located at the leading edge of migrating cells within 5 min after stimulation with a chemoattractant, N-formyl-methionine-leucine-phenylalanine. These membrane translocations also depend on PtdIns(3,4,5)P3 as both the PI3-kinase inhibitor LY294002 and mutations in the TH1 domain completely blocked the translocation. We also confirmed the localization for endogenous myosin IF using immunofluorescence with anti-myosin IF antibodies in HL-60 cells ().
Figure 2. Localization of endogenous myosin IF in HL-60 cells. HL-60 cells were stimulated with N-formyl-methionine-leucine-phenylalanine and subjected to immunofluorescence with anti-myosin IF antibodies (M-41, Santa Cruz). Myosin IF is located at the leading edge.
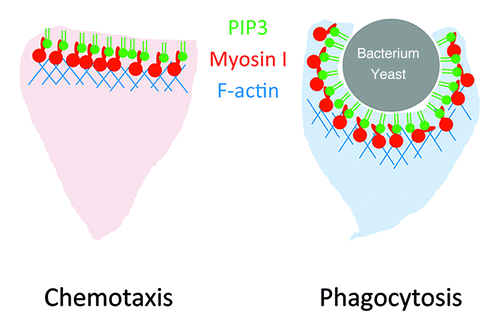
In addition to chemotaxis, we found that PtdIns(3,4,5)P3-binding myosin Is are also important for phagocytosis.Citation20 Like chemotaxis, PtdIns(3,4,5)P3 is generated at phagocytic cups and likely rearranges the actin cytoskeleton to engulf bacteria and yeast cells. We showed that PtdIns(3,4,5)P3-binding myosin Is are recruited to phagocytic cups in a PtdIns(3,4,5)P3-dependent manner and required for completion of phagocytosis. Mutations in the TH1 domain inhibited myosin I's recruitment to phagocytic cups and phagocytosis.
In chemotaxis and phagocytosis, PtdIns(3,4,5)P3 serves as a conserved signaling molecule which controls the actin cytoskeleton. Although additional, parallel signaling pathways exist, our recent findings and other previous studies suggest that myosin I plays important roles in these dynamic processes (). Considering that only a subset of myosin I binds to PtdIns(3,4,5)P3, different myosin I molecules are likely regulated by different mechanisms and act on distinct steps in intracellular signaling and actin cytoskeleton reorganization. It is of great interest to decipher how myosin I functions in dynamics of biological membranes and the cytoskeleton.
Acknowledgments
We thank H. Sesaki for critically reading the manuscript. This work was supported by a grant from NIH (GM084015).
References
- Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol 2001; 2:123 - 8; http://dx.doi.org/10.1038/84219; PMID: 11175804
- Murphy PM. Chemokines and the molecular basis of cancer metastasis. N Engl J Med 2001; 345:833 - 5; http://dx.doi.org/10.1056/NEJM200109133451113; PMID: 11556308
- Braunersreuther V, Mach F. Leukocyte recruitment in atherosclerosis: potential targets for therapeutic approaches?. Cell Mol Life Sci 2006; 63:2079 - 88; http://dx.doi.org/10.1007/s00018-006-6127-2; PMID: 16952057
- Mrass P, Weninger W. Immune cell migration as a means to control immune privilege: lessons from the CNS and tumors. Immunol Rev 2006; 213:195 - 212; http://dx.doi.org/10.1111/j.1600-065X.2006.00433.x; PMID: 16972905
- Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia 2007; 12:143 - 52; http://dx.doi.org/10.1007/s10911-007-9046-4; PMID: 17557195
- Affolter M, Weijer CJ. Signaling to cytoskeletal dynamics during chemotaxis. Dev Cell 2005; 9:19 - 34; http://dx.doi.org/10.1016/j.devcel.2005.06.003; PMID: 15992538
- Rørth P. Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev Cell 2011; 20:9 - 18; http://dx.doi.org/10.1016/j.devcel.2010.12.014; PMID: 21238921
- Bagorda A, Parent CA. Eukaryotic chemotaxis at a glance. J Cell Sci 2008; 121:2621 - 4; http://dx.doi.org/10.1242/jcs.018077; PMID: 18685153
- Nishio M, Watanabe K, Sasaki J, Taya C, Takasuga S, Iizuka R, et al. Control of cell polarity and motility by the PtdIns(3,4,5)P3 phosphatase SHIP1. Nat Cell Biol 2007; 9:36 - 44; http://dx.doi.org/10.1038/ncb1515; PMID: 17173042
- Subramanian KK, Jia Y, Zhu D, Simms BT, Jo H, Hattori H, et al. Tumor suppressor PTEN is a physiologic suppressor of chemoattractant-mediated neutrophil functions. Blood 2007; 109:4028 - 37; http://dx.doi.org/10.1182/blood-2006-10-055319; PMID: 17202315
- Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 2002; 109:599 - 610; http://dx.doi.org/10.1016/S0092-8674(02)00745-6; PMID: 12062103
- Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 2002; 109:611 - 23; http://dx.doi.org/10.1016/S0092-8674(02)00755-9; PMID: 12062104
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell 1998; 95:81 - 91; http://dx.doi.org/10.1016/S0092-8674(00)81784-5; PMID: 9778249
- Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev Cell 2002; 3:469 - 78; http://dx.doi.org/10.1016/S1534-5807(02)00292-7; PMID: 12408799
- Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem 2004; 279:16606 - 13; http://dx.doi.org/10.1074/jbc.M312098200; PMID: 14764604
- Meili R, Ellsworth C, Lee S, Reddy TB, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J 1999; 18:2092 - 105; http://dx.doi.org/10.1093/emboj/18.8.2092; PMID: 10205164
- Stephens L, Milne L, Hawkins P. Moving towards a better understanding of chemotaxis. Current biology: CB 2008; 18:R485-94.
- King JS, Insall RH. Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol 2009; 19:523 - 30; http://dx.doi.org/10.1016/j.tcb.2009.07.004; PMID: 19733079
- Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci U S A 2010; 107:11829 - 34; http://dx.doi.org/10.1073/pnas.1006153107; PMID: 20547830
- Chen CL, Wang Y, Sesaki H, Iijima M, Myosin I. Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci Signal 2012; 5:ra10; http://dx.doi.org/10.1126/scisignal.2002446; PMID: 22296834
- de la Roche MA, Côté GP. Regulation of Dictyostelium myosin I and II. Biochim Biophys Acta 2001; 1525:245 - 61; http://dx.doi.org/10.1016/S0304-4165(01)00110-6; PMID: 11257438
- Kim SV, Flavell RA. Myosin I: from yeast to human. Cell Mol Life Sci 2008; 65:2128 - 37; http://dx.doi.org/10.1007/s00018-008-7435-5; PMID: 18344022
- McConnell RE, Tyska MJ. Leveraging the membrane—cytoskeleton interface with myosin-1. Trends Cell Biol 2010; 20:418 - 26; http://dx.doi.org/10.1016/j.tcb.2010.04.004; PMID: 20471271
- Krugmann S, Anderson KE, Ridley SH, Risso N, McGregor A, Coadwell J, et al. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol Cell 2002; 9:95 - 108; http://dx.doi.org/10.1016/S1097-2765(02)00434-3; PMID: 11804589