Abstract
Eukaryotic transcriptome analyses have revealed that many transcripts are non-coding RNAs (ncRNAs). In addition, most relatively large (~several kb) polyadenylated mRNA type transcripts are transcribed from regions harboring little coding potential. However the role of such mRNA type long ncRNAs (mlonRNAs) is mostly unknown and has been a matter of debate. Recently, we showed that cascade of RNA polymerase II (RNAPII)-mediated transcriptional initiation of mlonRNA causes stepwise disruption of local chromatin array at the fission yeast Schizosaccharomyces pombe fbp1+ promoter region. Here, we hypothesize that RNAPII transcription of mlonRNA disrupt chromatin array possibly collaborating with histone acetylation mechanism. In addition, conserved action of Atf1, a transcriptional activator, and Tup11-Tup12 corepressors along mlonRNA transcription mediated chromatin regulation is suggested. This idea provides new insight into the biological meaning of mlonRNAs found in various eukaryotes.
Within a tiny nucleus, chromosomal DNA is compacted as a chromatin structure. The fundamental unit of chromatin is the nucleosome, consisting of histones wrapped with genomic DNA. The chromatin structure plays important roles in the expression and inheritance of genetic information in all eukaryotes. However, such chromatin compaction inhibits many DNA-related reactions, such as transcription, replication, DNA damage repair and recombination, by preventing the access of transacting DNA-binding factors to the DNA substrates.Citation1 Therefore, proper regulation of the chromatin structure is vitally important for the homeostasis of biological systems. The posttranslational modification of histones including acetylation and various chromatin remodeling complexes are known to regulate chromatin structure.Citation2–Citation4 In histone acetylation, a histone acetyltransferases (HATs) and deacetylases (HDACs) add and remove acetyl groups, respectively. Increased acetylation is usually associated with derepressed chromatin configuration.Citation5
Atf1, a CREB/ATF-type heterodimeric basic leucine zipper protein participates in the alteration of chromatin configuration into open structure and thereby roles in the transcriptional induction of some stress genes and activation of some set of meiotic recombination hotspot.Citation6–Citation8 In contrast Tup11 and Tup12, Groucho-like global corepressors repress the chromatin remodeling and thereby suppressing the transcriptional activation.Citation8–Citation11
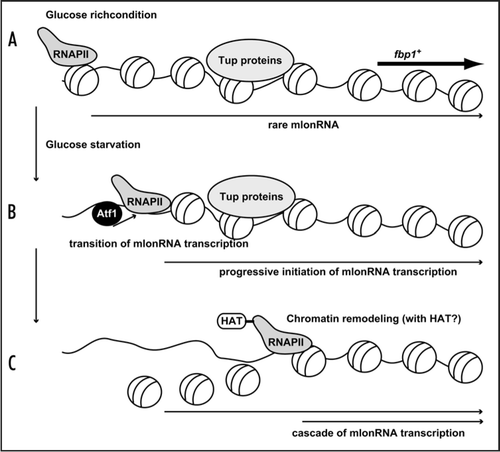
The fission yeast S. pombe fbp1+ gene is strictly regulated by glucose repression over a range of greater than 100-fold.Citation12 To study how such strict regulation is established, we have studied the chromatin regulation in fbp1+ promoter. We discovered a transient and cascaded transcriptional initiation of mRNA-type long ncRNA (mlonRNA) passing through the fbp1+ upstream region during the course of starvation-induced derepression.Citation13 In the course of cascaded fbp1+ mlonRNA transcription, RNA polymerase II (RNAPII) translocates from far upstream to eventual transcriptional start site in the fbp1+ promoter.Citation13 Interestingly, chromatin structure progressively convert into open configuration from far upstream from the fbp1+ promoter and is induced in a stepwise manner 5′ to 3′ toward the fbp1+ promoter.Citation13 Noteworthy, this chromatin-remodeling event is coupled with transcriptional transition as well as the translocation of RNAPII along fbp1+ upstream region. Moreover, we showed this cascaded mlonRNA transcription is vital for the chromatin remodeling event, since arrest of mlonRNA transcription by the insertion of a transcription terminator abolishes the progressive chromatin alteration.Citation13 We therefore concluded that RNAPII transcription of mlonRNA disrupts chromatin array within its passed tract. Since we detected transient and cascaded histone acetylation along fbp1+ upstream region(our unpublished results), it is possible that RNAPII travels along non-coding fbp1+ upstream region and disrupts its passed tract collaborating with HAT activity (). Furthermore, we demonstrated that Atf1 is required for the progressive mlonRNA transition as well as the stepwise chromatin-remodeling event. However, concomitant loss of Tup11 and Tup12 corepressors in atf1Δ cells rescues massive transcription of fbp1+ from TATA box without recovering mlonRNA transition. Hence, a possible scenario is that Atf1 activates progressive mlonRNA initiations and thereby overcomes the repressive role of the Tup proteins ().
As a similar chromatin alteration event, we previously reported the coupling of chromatin alteration and shift of transcription initiation site at meiotic recombination hotspot ade6-M26, in which a non-sense mutation simultaneously creates cyclic AMP responsible element (CRE) like sequence that is responsible for the hotspot activity.Citation14 Noteworthy, in the regulation of chromatin structure at ade6-M26 site, Atf1 and Tup proteins roles in the same manner as in fbp1+ promoter.Citation6,Citation9 These similarities led us to speculate that mlonRNA transcription regulates chromatin structure possibly collaborating with Atf1 and Tup proteins. Such sophisticated chromatin regulation system consisting mlonRNA transcription, Atf1 and Tup proteins could be important also in higher eukaryotes, because the system consisting of MAPK pathways-Atf1 transcription factor and Tup proteins are highly conserved and many of ncRNAs of unknown function are found in various eukaryotes.Citation15–Citation19 While the biological meaning of such nc-RNAs has been mostly unknown so far, the ‘mlonRNA-coupled chromatin regulation’ presented here provides important clues to understand ncRNA transcription found in various eukaryotes.
Figures and Tables
Figure 1 A model representing mlonRNA transcription disrupts chromatin array. (A) In glucose rich condition, rare mlonRNA is transcribed from a site far upstream from the authentic fbp1+ promoter, but does not initiate the robust activation of fbp1+ transcription at the promoter due to the Tup-dependent repressive chromatin structure. (B) Upon glucose starvation, Atf1 binds upper binding site (carrying CRE sequence). Atf1 activates progressive mlonRNA initiations, and this mlonRNA transcription overcomes the repressive role of the Tup proteins. (C) RNAPII traveling along upper fbp1+ region disrupts chromatin array possibly collaborating with HAT activity.
Addendum to:
References
- Wolffe A. Chromatin: Structure and Function 1997; 3rd Ed San Diego, CA Academic Press
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle and developmentally regulated promoter. Cell 1999; 97:299 - 311
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFNbeta promoter. Cell 2000; 103:667 - 678
- Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev 1999; 13:1412 - 1421
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol 1998; 8:193 - 197
- Yamada T, Mizuno KI, Hirota K, Kon N, Wahls WP, Hartsuiker E, et al. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J 2004; 23:1792 - 1803
- Hirota K, Steiner WW, Shibata T, Ohta K. Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast. Eukaryot Cell 2007; 6:2072 - 2080
- Hirota K, Hasemi T, Yamada T, Mizuno KI, Hoffman CS, Shibata T, et al. Fission yeast global repressors regulate the specificity of chromatin alteration in response to distinct environmental stresses. Nucl Acids Res 2004; 32:855 - 862
- Hirota K, Hoffman CS, Shibata T, Ohta K. Fission yeast Tup1-like repressors eepress chromatin remodeling at the fbp1+ promoter and the ade6-M26 recombination hotspot. Genetics 2003; 165:505 - 515
- Hirota K, Hoffman CS, Ohta K. Reciprocal nuclear shuttling of two antagonizing Zn finger proteins modulates Tup family corepressor function to repress chromatin remodeling. Eukaryot Cell 2006; 5:1980 - 1989
- Janoo RT, Neely LA, Braun BR, Whitehall SK, Hoffman CS. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 2001; 157:1205 - 1215
- Hoffman CS, Winston F. A transcriptionally regulated expression vector for the fission yeast Schizosaccharomyces pombe. Gene 1989; 84:473 - 479
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodeling by a cascade of transcription initiation of non-coding RNAs. Nature 2008; 456:130 - 134
- Hirota K, Mizuno KI, Shibata T, Ohta K. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol Biol Cell 2008; 19:1162 - 1173
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005; 308:1149 - 1154
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 2004; 116:499 - 509
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science 2005; 309:1559 - 1563
- Hayashizaki Y, Carninci P. Genome Network and FANTOM3: assessing the complexity of the transcriptome. PLoS Genet 2006; 2:63
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 2008; 453:1239 - 1243