Abstract
The plasma membrane of mammalian cells is composed of a great variety of different lipids which are laterally organized into lipid domains. The segregation of lipids into domains has been studied in great detail in vesicles but domain formation of lipids in the plasma membrane of live cells is still unclear. We have previously used fluorescence lifetime imaging microscopy to study the colocalization of the receptor for EGF with the ganglioside GM1 and the GPI-anchored green fluorescent protein. Here we have used this technology to study the effect of EGF on the organization of GM1 in the plasma membrane. Our data show that stimulation of the cell with EGF induces rapidly a strong increase in colocalization of GM1 molecules, suggesting the formation of large lipid domains. These results support the notion that activation of EGFR signaling may result in the formation of signaling platforms.
The plasma membrane functions as a barrier separating the cells interior from the extracellular environment. In addition to its barrier function the plasma membrane also functions as a recruitment site for signaling processes, for instance by the binding of inositol lipids to PH-domain containing signaling molecules as the serine/threonine kinase Akt.Citation1,Citation2 The membrane is composed of a large variety of different lipids of which some have a clear structural function (phosphatidylcholine), while others can act as first and second messenger in signal transduction phosphatidylinositol.Citation3 A particular property of the plasma membrane is the relative high concentration of sphingolipids in the outer leaflet and the non-polar sterol cholesterol in both leaflets of the membrane. Biochemical and biophysical studies have indicated that mixtures of these three types of lipids can form particular domains of either phospholipids or sphingolipids stabilized by cholesterol.Citation4 Questions that remain, however, are whether such domains exist in the plasma membrane of living cells and moreover, how this behavior of membrane molecules relates to their function.
To study these questions we have investigated the colocalization of the receptor for the growth factor EGF with proteins and lipids that are typical for lipid rafts in more detail. From biochemical studies it was already concluded that EGFR is present in parts of the plasma membrane that are resistant to extraction by Triton X-100, called detergent-resistant membrane domains (DRMs).Citation5 These domains, isolated as light buoyant fractions on a density gradient, are thought to represent clusters of lipids carrying saturated fatty acids, and cholesterol. In model membranes, the saturation of the fatty acids allows the tight packing with cholesterol, which drives phase separation of these liquid ordered (LO) domains from the remaining liquid disordered (LD) bilayer. Partitioning of certain proteins and lipids into these ‘lipid rafts’ can therefore enable their mutual interaction. A lipid class that co-purifies with this fraction are the gangliosides, which are an important factor in the regulation of EGFR. Gangliosides are characterized by their glycan moiety including minimally one sialic acid residue. Recently, primary fibroblasts were isolated from patients suffering from an autosomal recessive mutation in GM3 synthase, an essential enzyme in ganglioside synthesis, which caused a 93% reduction of cellular ganglioside levels.Citation6 These cells display a highly reduced EGF binding although EGFR levels were unaffected. Also EGF-induced proliferation and migration were reduced, which correlated with reduced EGFR phosphorylation and Rho/Rac1 activation. How gangliosides exert their influence on the EGFR is far from clear. It has been suggested that the glycan head-groups can differentially affect the conformation of the ectodomain, preventing or enhancing favorable conformations for signaling. This view is supported by the ability of EGFR to directly bind the ganglioside glycans, depending on the glycosylation of the receptor.Citation7
Although the biochemical analysis of lipid rafts in vitro has yielded a large amount of information, application of this approach to the cellular situation suffers from a number of limitations that frustrate reliable interpretations. Detergent extraction has the risk of artificial clustering of membrane components, gives no information on the spatial distribution of domains, and does not discriminate between different domains in a heterogeneous population. Moreover, the sensitivity of proteins for extraction varies with the type of detergent used (TX-100, Brij-98 or 96, Tween-20, octylglucoside), suggesting that the discrimination between LO and LD is more complex.Citation5,Citation8
To study the orientation of the EGFR in the plasma membrane we have previously set up a microscopical approach.Citation9 The putative submicron scale of membrane domains cannot directly be visualized in the light microscope, because the optical resolution limit, determined by the wavelength of light, does not allow discrimination of separate objects smaller than ∼200 nm. The colocalization of the EGFR and components of lipid rafts was therefore investigated by Förster resonance energy transfer (FRET).Citation10 FRET is a process in which energy is transferred from an excited fluorescent donor molecule to a fluorescent acceptor probe. It can be used to detect the 5–10 nm proximity of molecules. The extent to which FRET takes place is measured by means of fluorescence lifetime imaging microscopy (FLIM); the occurrence of FRET results in a reduction of the donor's fluorescence lifetime.
An important aspect in this approach concerns the conjugation of the fluorescent probes to the molecules of interest, i.e., EGFR and GM1. For the labeling of GM1 we have used the B-subunit of cholera toxin (CTB) directly conjugated to an Alexa Fluor probe. As a marker for the EGFR we have used nanobodies which are the single chain epitope-binding subunits from the heavy chain only antibodies from Llama glama.Citation11 Besides their small size (∼15 kDa), nanobodies can be selected using phage display for their high affinity binding to the ectodomain of the EGFR.Citation11 Likewise, we generated a number of anti-EGFR nanobodies that neither compete for EGF-binding nor activate the EGFR, and which binding characteristics are not affected by conjugation of the fluorescent probe.Citation9 Our results demonstrated the presence of GM1 in the proximity of the EGFR. However, another typical lipid raft resident molecule, a GPI-anchored green fluorescent protein did not colocalize in the resting cell with the EGFR. Interestingly, the colocalization of GM1 with GPI-GFP appeared to be cholesterol-dependent while the colocalization of the EGFR with GM1 was independent of cholesterol. These results suggest the presence of different classes of lipid raft in the resting cell.
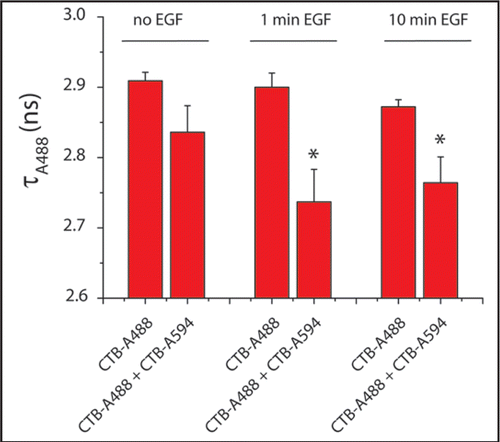
Stimulation of EGF signaling did not affect the colocalization of EGFR with GM1. By contrast, EGFR activation induced the colocalization of the EGFR with GPI-GFP, suggesting the coalescence of different lipid rafts upon receptor activation. To further analyze the effect of EGF on the organization of lipid rafts we analyzed the nanoscale colocalization of GM1 during EGF signaling. FRET efficiency between differentially labeled CTB molecules was analyzed in time by measuring the fluorescence lifetime of the donor probe (). In the resting cell no significant colocalization was observed between CTB-A488 (donor) and CTB-A594 (acceptor). Already after 1 minute of EGFR activation a significant increase in FRET was observed between the CTB molecules demonstrating an increase in proximity of GM1 lipids. Such data suggest that EGF induces the formation of larger lipid rafts, which may lead to alterations in the inner leaflet of the plasma membrane as well. Evidence for such transbilayer effects have been demonstrated in vitro using asymetric planar bilayers.Citation12 Considering the more effective activation of the PH-containing serine/threonine kinase Akt within lipid microdomains,Citation1,Citation2 the EGF-induced changes in the organization of the plasma membrane may result in the formation of signaling platforms, which represent highly efficient signaling sites in the plasma membrane.
Abbreviations
| A488 | = | alexa fluor 488 |
| CTB | = | cholera toxin B-subunit |
| FLIM | = | fluorescence lifetime imaging microscopy |
| FRET | = | Förster resonance energy transfer |
Figures and Tables
Figure 1 EGF stimulation induces coalescence of GM1-containing rafts. Serum-starved HER14 cells (NIH 3T3 cells stably expressing human EGFR), were labeled with 1 µg/ml cholera toxin B-subunit conjugated to Alexa Fluor 488 (CTB-A488) and/or Alexa Fluor 594 (CTB-A594) on ice. After 1 hour, the cells were recovered to 37°C and stimulated with 20 ng/ml EGF for 1 or 10 minutes. The cells were fixed with 4% formaldehyde and the coverslips were embedded in mowiol. The lifetimes of Alexa 488 were determined by FLIM analysis as described previously.Citation9 Histograms show the average lifetime values of four cells per condition (*p < 0.05).
Acknowledgements
This work was supported by the ‘From Molecule to Cell’ program from the Dutch scientific organisation (NWO-ALW, grant 805.47.084) to Erik Hofman and Arjen Bader.
Addendum to:
References
- Gao X, Zhang J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol Biol Cell 2008; 19:4366 - 4373
- Lasserre R, Guo XJ, Conchonaud F, Hamon Y, Hawchar O, Bernard AM, et al. Raft nano-domains contribute to Akt/PKB plasma membrane recruitment and activation. Nat Chem Biol 2008; 4:538 - 547
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008; 9:112 - 124
- Jacobson K, Mouritsen OG, Anderson RG. Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol 2007; 9:7 - 14
- Pike LJ, Han X, Gross RW. Epidermal growth factor receptors are localized to lipid rafts that contain a balance of inner and outer leaflet lipids: a shotgun lipidomics study. J Biol Chem 2005; 280:26796 - 26804
- Liu Y, Su Y, Shevchuk NA, Wiznitzer M, Epifano O, Ladisch S. Ganglioside depletion and EGF Responses of Human GM3 Synthase Deficient Fibroblasts. Glycobiology 2008; In press
- Miljan EA, Meuillet EJ, Mania-Farnell B, George D, Yamamoto H, Simon HG, Bremer EG. Interaction of the extracellular domain of the epidermal growth factor receptor with gangliosides. J Biol Chem 2002; 277:10108 - 10113
- Delaunay J-L, Breton M, Trugnan G, Maurice M. Differential solubilization of inner plasma membrane leaflet components by Lubrol WX and Triton X-100. Biochim Biophys Acta 2008; 1778:105
- Hofman EG, Ruonala MO, Bader AN, van den Heuvel D, Voortman J, Roovers RC, et al. EGF induces coalescence of different lipid rafts. J Cell Sci 2008; 121:2519 - 2528
- Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol 2003; 21:1387 - 1395
- Roovers R, Laeremans T, Huang L, De Taeye S, Verkleij A, Revets H, et al. Efficient inhibition of EGFR signalling and of tumour growth by antagonistic anti-EGFR Nanobodies. Cancer Immunol Immunother 2006; 56:303 - 317
- Kiessling V, Crane JM, Tamm LK. Transbilayer effects of raft-like lipid domains in asymmetric planar bilayers measured by single molecule tracking. Biophys J 2006; 91:3313 - 3326