Abstract
Protein synthesis in eukaryotes initiates with binding of the multisubunit translation initiation complex eIF4F. This complex contains eIF4E, eIF4A and eIF4G. eIF4E directly interacts with the cap structure, eIF4A is an RNA helicase and eIF4G acts as a scaffold for the complex. eIF4G contains the binding sites for both the subunits i.e. eIF4A and eIF4E and it also interacts with poly(A)-binding protein (PABP). In present study we have identified and characterized the main components of the eIF4F complex i.e. eIF4E, eIF4A and eIF4G and PABP from Plasmodium falciparum. Molecular modeling of PfeIF4E, PfeIF4G and PfPABP confirms that they contain all the characteristic conserved structural features. We have annotated some of the genes of P. falciparum and as a result these studies demonstrate that the components of translation initiation complex are highly conserved. Therefore these studies will contribute to understand the basic biology and components of translation complex in P. falciparum.
Generally eukaryotic mRNAs contain a 5′ cap (m(7)GpppN) where N is any nucleotide and a 3′ poly(A) tail (50–300 nucleotides), both of which are essential for efficient mRNA translation and are specifically recognized by the proteins. Eukaryotic translation initiation is a multistage process, which involves several sequential steps performed by a multisubunit protein complex termed as eukaryotic translation initiation factor 4F (eIF4F).Citation1,Citation2 eIF4F is composed of multiple translation factors, which assemble at the monomethylated cap present on the 5′-end of the mRNA to promote the recruitment of the ribosome. eIF4F mainly consists of three subunits, eIF4A (eukaryotic translation initiation factor 4A), an ATP-dependent RNA helicase and a member of the DEAD-box protein family (DEAD corresponds to Asp-Glu-Ala-Asp), eIF4E (eukaryotic translation initiation factor 4E) which binds to the 5′ cap structure of the mRNA and eIF4G (eukaryotic translation initiation factor 4G) that acts as a scaffold for the complex with binding sites for both eIF4A and eIF4E. eIF4G is also responsible for the circularization of the mRNA via interaction with poly(A) binding protein (PABP).Citation3 The 5′ cap and the 3′ poly(A) tail of eukaryotic mRNAs cooperate to synergistically increase translation.
The functions of the components of eIF4F complex have been well established by studies in various systems. eIF4A is believed to be involved in the unwinding of the inhibitory secondary structures present in the 5′ untranslated region of the mRNA and its maintenance in a single stranded form to facilitate the binding and scanning of the ribosomes to the initiator AUG codon.Citation4,Citation5 eIF4E mediates cap identification during initiation and is absolutely necessary for cap-dependent translation.Citation6 Most organisms contain a single gene, which encodes eIF4E but some organisms contain more than one eIF4E gene, which belong to different families.Citation7 eIF4G is an adapter protein with a modular structure, which is required to mediate the ribosome recruitment via the cap structure and poly(A) tail.Citation1 Mammals possess two functional isoforms of eIF4G eIF4GI and eIF4GII.Citation2 The binding sites for numerous interaction partners of eIF4G have been determined by mutation and deletion analysis and it has been suggested that eIF4G provides multiple contact points for interaction with other components of the initiation complex.Citation2 It has been shown that mammalian eIF4G contains two interaction domains for eIF4A, a C-terminal domain and a central domain and the interaction of eIF4A with the middle region of eIF4GI is necessary for translation, whereas the interaction of eIF4A with the C-terminal region plays a modulatory role.Citation8,Citation9 PABPs usually bind poly(A) using one or more globular domains containing RNA-recognition motifs (RRMs) and therefore help in stabilization of mRNAs. PABPs are involved in interaction of the poly(A) tail with the translation initiation complex and simultaneous binding of eIF4E and PABP to eIF4G is essential for efficient translation.Citation2,Citation3,Citation10 The highly conserved PABP polypeptides are found only in eukaryotes; single-celled eukaryotes each contains 1–2 PABP genes, while humans have five and Arabidopsis has up to eight PABP genes.Citation3,Citation11 Several molecules of PABP cover the entire length of the poly(A) tail of the mRNA and this interaction occurs via RRMs of PABP.Citation3,Citation11
Malaria is a major parasitic infection caused by the protozoan parasites of the genus Plasmodium and Plasmodium falciparum causes the most virulent form of malaria.Citation12 In order to understand the basic biology and specially the translation process in this protozoan parasite we have bioinformatically characterized various components of the eIF4F complex from P. falciparum. In previous studies we have reported the detailed characterization of an eIF4A homologue designated as PfH45 from P. falciparum.Citation13,Citation14 In the present manuscript we present characterization of the other principal components of the eIF4F complex such as eIF4E, eIF4G and PABP from P. falciparum. Our studies reveal that P. falciparum contains only one gene each encoding eIF4E, eIF4G and PABP. The bioinformatics characterization of these proteins reveals that PfeIF4E, PfeIF4G and PfPABP all contain the characteristics features. PfPABP also contains the characteristic RRMs. A three-dimensional homology model of the middle domain of PfeIF4G shows that there is sufficient conservation to human eIF4G to make the models virtually super-imposable. The studies reported here show that despite the insignificant homology of PfeIF4G at the sequence level its structure is highly conserved. These studies will make an important contribution towards understanding the process of translation in the parasite.
Sequence Analysis and Computer Based Prediction of Structure of eIF4E Homologue from P. falciparum
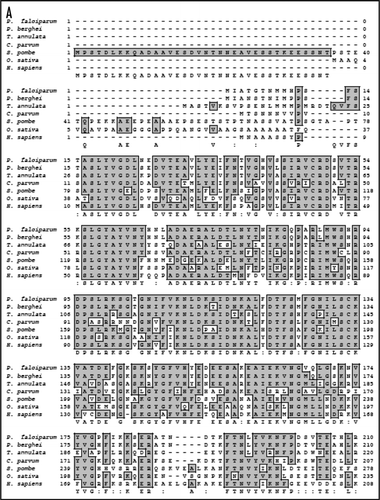
The search of the P. falciparum genome using eIF4E as query revealed that only one homologue of eIF4E is present in the genome. The 684 base-pair open reading frame of P. falciparum eIF4E (PfeIF4E) was analyzed. The deduced amino acid sequence of PfeIF4E revealed a protein consisting of 227 amino acids with a predicted molecular mass of approximately 27 kDa and it is a basic protein with a calculated isoelectric point of ∼8.4. The sequence analysis confirmed the presence of characteristic motifs of this family including the conserved tryptophan residues.Citation7 The ‘PlasmoDB’ (http://www.plasmodb.org) entry number for this gene is PFC0635c and it contains no introns. A multiple alignment of amino-acid sequence homology search using NCBI database revealed that PfeIF4E aligned contiguously and showed highest homology with its counterparts from Plasmodium berghei (∼92%) and ∼19–40% homology with eIF4E from other sources ( and B).
The 3 dimensional structures of the yeast eIF4E determined by NMR and mouse and human eIF4E proteins determined by X-ray crystallography have been reported.Citation15–Citation18 The murine protein has a cupped hand shape and contains eight-stranded anti-parallel β-sheets, backed by three α-helices on its convex side.Citation16 A thorough examination of the alignment of PfeIF4E revealed that the cap-binding pocket is highly conserved. A comparison of the amino acids of human eIF4E and PfeIF4E is shown in . The interactions mediated by the amino acids required for specific binding to the 7-methyl-GDP are taken over by the amino acids Trp37, Asp102, Trp115, Glu116, Lys170, Arg180, His125 and Trp184 in PfE27,Citation16 (, marked by red asterisk). Similarly the alignment showed that the amino acids His37, Pro38, Val69, Trp73, Leu128 and Leu135 of mammalian eIF4E,Citation19 which are responsible for binding to eIF4G, are replaced by the amino acids Leu31, Leu32, Val67, Trp71, Asn145 and Leu149 in PfeIF4E (, marked by blue asterisk). For the location of the binding pocket on PfeIF4E, the complete protein sequence was submitted to the JIGsaw program (http://www.bmm.icnet.uk/servers/3djigsaw/).Citation20–Citation22 A highly similar and superimposable model based on the crystal structure of human eIF4E was obtainedCitation16 [ (i)]. In these models the amino acids responsible for the binding of eIF4G have been marked using the VMD software (www.ks.uiuc.edu) and are shown in yellow color [ (ii) and (iii)]. These results further show that the binding pocket is present on the similar planes in PfeIF4E and human eIF4E.
Sequence Analysis and Computer Based Prediction of Structure of eIF4G Homologue from P. falciparum
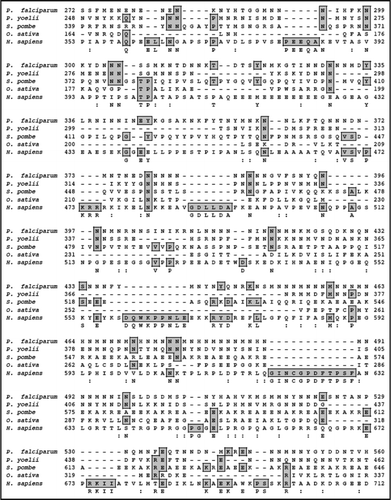
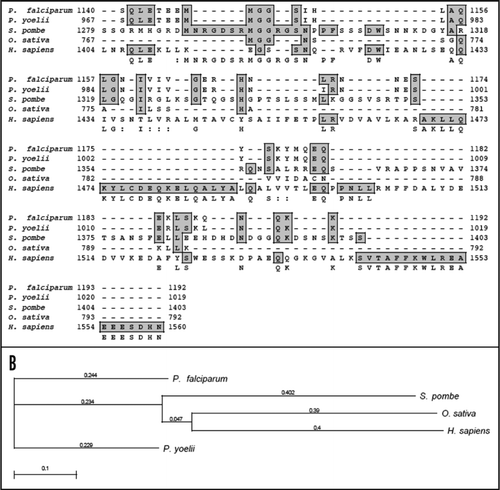
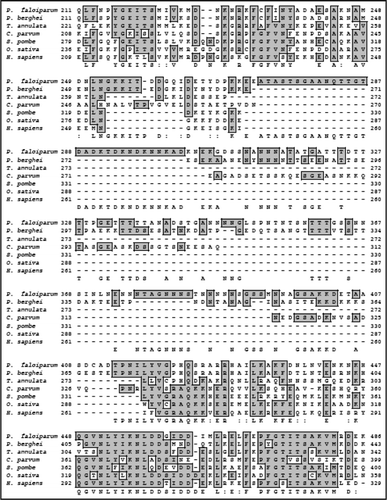
On simple text-based data mining of PlasmoDB with the text term ‘MIF4G’ five genes are reported. These are PF11_0086 (hypothetical protein of 3334 amino acid in PlasmoDB), PFI1265w (hypothetical protein of 1754 amino acid in PlasmoDB), PFL1855w (putative cell cycle control protein of 967 amino acid in PlasmoDB), PF14_0113 (hypothetical protein of 943 amino acid in PlasmoDB) and MAL13P1.63 (asparagine-rich protein of 1192 amino acid in PlasmoDB). The detailed analysis of all of these proteins revealed that these contain MIF4G domain but all the others except MAL13P1.63 did not reveal any detectable homology to known proteins from other organisms. The detailed sequence analysis of MAL13P1.63 (∼139 kDa protein) confirmed the presence of characteristic motifs of eIF4G. A multiple alignment of amino-acid sequence homology search using NCBI database revealed that PfeIF4G aligned contiguously and showed highest homology with its counterparts from Plasmodium yoelii (∼49%) and ∼6–13% homology with eIF4G from other sources ( and B).
Since PfeIF4G contained insignificant homology at amino acid level therefore to check the homology at structural level, the complete protein sequence of PfeIF4G was submitted to the 3D-JIGSAW program and a three-dimensional model was obtained, which was based on the X-ray structure of the template human eIF4G.Citation23 On careful analysis it was observed that this model was built by using only the phylogenetically conserved region i.e., amino acid 888 to amino acid 1125 in PfeIF4G ( (i)). It is interesting to note that this domain is composed of ten α helices arranged as five anti-parallel α helical pairs or HEAT repeats (named for Huntington, Elongation factor 3, A subunit of protein phosphatase 2A [PP2A], and Target of rapamycin).Citation24 Similarly using the complete protein sequence of human eIF4G (AF104913) a three dimensional model was obtained [ (ii)]. This model spanned from amino acid 712 to 944 of human eIF4G. The alignment of amino acid 888 to 1125 of PfeIF4G and amino acid 712 to 944 of human eIF4G showed only 22% similarity and 27% identity (). But it is interesting to note that the two structures of these regions are highly similar and largely super-imposable [ (iii)].
Sequence Analysis and Computer Based Prediction of Structure of PABP Homologue from P. falciparum
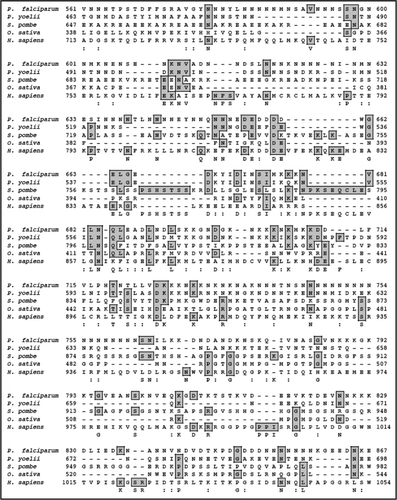
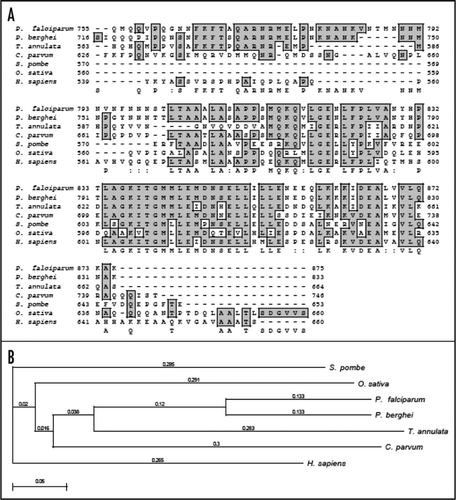
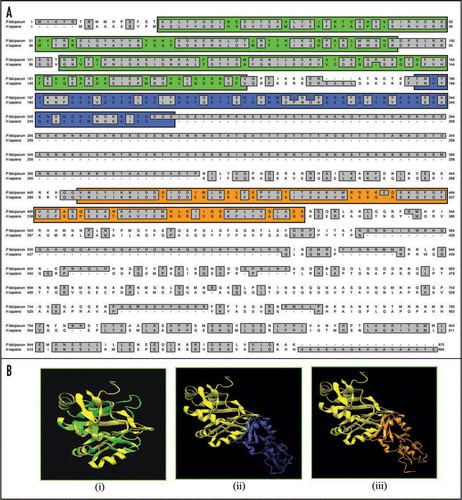
On simple text-based data mining of PlasmoDB with the text term ‘poly A binding protein’ two genes are reported. These are MAL13P1.303 (putative polyadenylate binding protein of 414 amino acids) and PFL1170w (putative polyadenylate binding protein of 875 amino acids). MAL13P1.303 is small in size to be a bonafide PABP. The detailed sequence analysis of PFL1170w revealed a protein with a predicted molecular mass of ∼97 kDa and it is a basic protein with a calculated isoelectric point of ∼8.9. A multiple alignment of amino-acid sequence homology search using NCBI database revealed that PFL1170w (PfPABP) aligned contiguously and showed highest homology with its counterparts from Plasmodium berghei (∼71%) and ∼29–37% homology with PABP from other sources (). It was observed that similar to other PABPs, PfPABP also contains four RRMs, which span from amino acid 16–94; 104–176; 193–261 and 450–527 respectively ().Citation11 It contains a few regions, which are Asn rich (amino acid 295–396; 617–799), Thr rich (amino acid 317–362) and Gln rich (amino acid 557–761) (). The presence of homorepeats is a characteristic feature of proteins from P. falciparum and the most common is asparagines followed by lysines and glutamine.Citation25,Citation26 These inserted residues encode non-globular domains of unidentified function, which are extruded from the core of the protein and probably form surface exposed structure that consequently in most of the proteins have no effect on the functional folding of the protein.Citation25,Citation26
The complete protein sequence of PfPABP was submitted to the 3D-JIGSAW program and a three-dimensional model was obtained, which was based on the X-ray structure of the template human PABP. These two structures were not similar and not super-imposable (data not shown). The amino acid alignment of full-length human and PfPABP revealed a similarity of ∼14% and an identity of ∼33% and it further showed that PfPABP contains an insertion of ∼150 amino acids in between RRM3 and 4 (). It is interesting to note that only the modeled structure of PfPABP from amino acids 1–180 (RRM1 and 2) superimposes with that of human PABP [ (i)] whereas the structures of PfPABP from amino acids 1–264 (RRM1–3) and from amino acids 1–531 (RRM1–4) are not superimposable with human PABP [ (ii) and (iii) respectively].
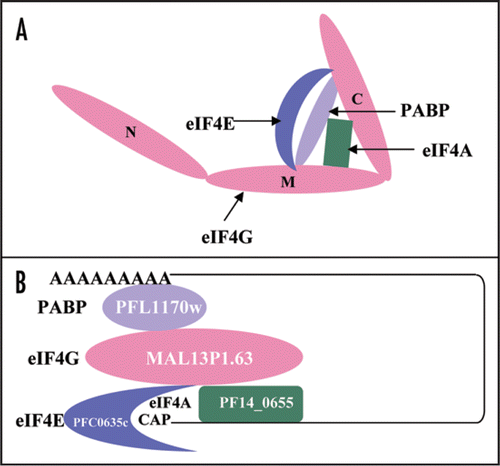
Our unpublished observations indicate that the middle and C-terminal domain of PfeIF4G is structurally conserved and is able to interact with PfH45, PfeIF4E and PfPABP but there was no detectable interaction between the PfeIF4G-N fragment and PfeIF4E or PfH45 or PfPABP. These interactions are shown schematically in and B shows the overall initiation complex of P. falciparum.
Protein synthesis is a complex process in eukaryotes and it requires numerous different macromolecules. The critical step of translation initiation is a coordinated action of a number of translation initiation factors. Most important of these is the heterotrimeric eIF4F complex, which is composed of eIF4A, eIF4E and the very large protein eIF4G, which mediates the interactions between eIF4F and other translation factors. Previously we have reported the characterization of an eIF4A homologue (PfH45) from P. falciparum and have shown that PfH45 is localized in the cytoplasm and is essential for parasite survival.Citation13 In the present study we have isolated and characterized the other components i.e., eIF4E and eIF4G of eIF4F complex and PABP from P. falciparum using the bioinformatics approach. Previously it has been reported that PfeIF4E binds to mRNAs in P. falciparum.Citation27
Previous studies have shown that the HEAT repeat proteins participate in a wide variety of cellular processes, which are dependent on accumulating large multiprotein complexes.Citation23,Citation24 The overall similarity between human eIF4G and PfeIF4G is not very high (only ∼12%) because it is well established that the HEAT repeat proteins do not share any absolutely conserved amino acids.Citation23 It has been reported that although Leishmania major contains five eIF4G homologues but most of them show similarity to the human sequence only at the level of central HEAT domain.Citation28 The comparison of human and P. falciparum eIF4G suggests that specifically the middle region of PfeIF4G might be involved in the interaction with PfH45 as reported for human eIF4G and eIF4A.Citation24 Due to the presence of HEAT domain, the conserved middle domain of PfeIF4G most likely serves as a central assembly platform for its translation initiation machinery. It has been reported previously that mammalian eIF4A binds to the central region of eIF4G via its HEAT domain and it also binds to its C-terminal domain.Citation29 Similarly in yeast it has been shown that eIF4G and eIF4A physically associate and this association is required for translation and is essential for yeast cell viability.Citation30 The binding site for eIF4E in human eIF4G has been reported previouslyCitation31,Citation32 and the alignment of the human and PfeIF4G shows that this site in PfeIF4G is between amino acid 464 to 473 and these amino acids are present in the middle domain of PfeIF4G.
It is interesting to note that P. falciparum contains only one PABP, which is localized in the cytoplasm and as reported earlier evolutionarily PfPABP is closer to PABP from L. major.Citation3 Multiple PABPs, both nuclear and cytoplasmic have been reported in human.Citation33 It has been reported previously that the amino acid sequences of the PABP-binding site in eIF4G are not conserved between yeastCitation31 and human.Citation32 The interaction between PABP and eIF4G is conserved in many species and this interaction can be imparted to RRM 1 and 2 of PABP.Citation33 Previously it has been reported that the interaction of eIF4G with PABP requires RRM1 and 2 in animal and yeast systemsCitation3,Citation32,Citation34 but in wheat PABP RRM1 was sufficient to interact with eIF4G.Citation35 The structural modeling of PfPABP suggests that the RRM1 and 2 are structurally conserved and PfPABP is also able to interact with the middle and C-terminal domain of PfeIF4G.
Overall in this study we have reported the bioinformatics characterization of important components of translation initiation complex eIF4F and PABP from P. falciparum. We have shown that the scaffold protein PfeIF4G contains the characteristic features for binding to PfeIF4E, PfeIF4A (PfH45) and PfPABP and thus provides the assembly platform for the components of the initiation complex. In order to control malaria, it is essential to understand the basic biology of malaria parasite. Therefore this study makes an important valuable contribution towards better understanding of the function of main components of one of the basic metabolic pathways, the translation, in the malaria parasite.
Figures and Tables
Figure 1 (A) Comparison of amino acid sequences of P. falciparum eIF-4E protein (PlasmoDB No. PFC0635c; GenBank accession No. EF043517) with other eIF-4E from Plasmodium berghei (XP_674865), Theileria annulata (CAI74251), Cryptosporidium parvum (CAD98650), Pisum sativum (ABG35118), Saccharomyces cerevisiae (NP_014502) and Homo sapiens (P06730). The accession numbers of the aligned sequences are written in brackets. (B) Phylogenetic guide tree using Clustal W program based on the multiple alignment. A value of 0.1 corresponds to a difference of 10% between two sequences.

Figure 2 (A) Amino acid alignment of P. falciparum eIF4E (PlasmoDB No. PFC0635c, GenBank accession number EF043517) and human eIF4E (GenBank accession number P06730). The amino acids responsible for 7-methyl GDP-binding and eIF4G binding are marked by red and blue asterisk respectively. (B) A three-dimensional model for PfeIF4E was created as described in text, which was based on the crystal structure of human eIF4E.Citation16 The structures have been displayed using molecular visualization program for displaying, animating and analyzing large biomolecule systems using 3-dimensional graphics and built-in scripting (VMD software www.ks.uiuc.edu). (i) Super imposed image of the structure of human (green) and P. falciparum (red) eIF4E The amino acids responsible for the binding of eIF4G in (ii) human and (iii) P. falciparum eIF4E have been marked and are shown in yellow in the models.
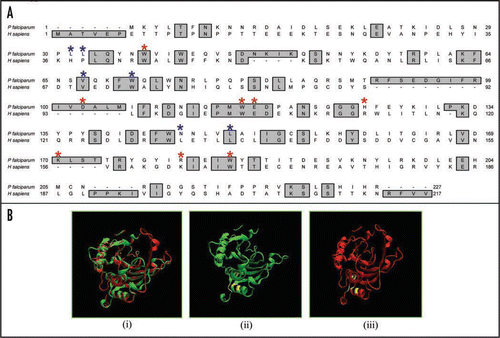
Figure 3 (A) Comparison of amino acid sequences of P. falciparum eIF-4G protein (PlasmoDB No. MAL13P1.63) with other eIF4G from Plasmodium yoelii (XP_727991), Schizosaccharomyces pombe (Q10475), Oryza sativa (AAO72569) and Homo sapiens (AF104913). The accession numbers of the aligned sequences are written in brackets. (B) Phylogenetic guide tree using Clustal W program based on the multiple alignment. A value of 0.1 corresponds to a difference of 10% between two sequences.
Figure 4 (A) Comparison of amino acid sequence (from amino acid 888-1125) of P. falciparum eIF4G protein (PlasmoDB no. MAL13P1.63) with eIF4G (from amino acid 712-944) of H. sapiens (GenBank accession number AF104913). (B) Three-dimensional models for (i) human eIF4G and (ii) P. falciparum eIF4G were created as described in text. The structures have been displayed using the same program as described in legend to . This model has been built by using only the phylogenetically conserved middle domain i.e., amino acid 888 to amino acid 1125 in PfeIF4G. A super-imposed image is shown in (iii).
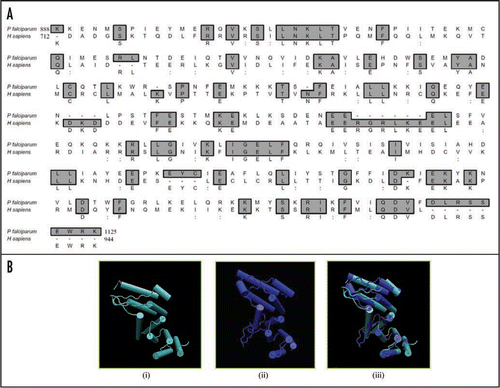
Figure 5 (A) Comparison of amino acid sequences of P. falciparum PABP protein (PlasmoDB No. PFL1170w; GenBank accession No. EF116593) with other PABP from Plasmodium berghei (XP_677383), Theileria annulata (CAI73150), Cryptosporidium parvum (CAD98589), Schizosaccharomyces pombe (AAA35320), Oryza sativa (XP_481529) and Homo sapiens (CAI12298). The accession numbers of the aligned sequences are written in brackets. (B) Phylogenetic guide tree using Clustal W program based on the multiple alignment. A value of 0.1 corresponds to a difference of 10% between two sequences.
Figure 6 (A) Comparison of amino acid sequence of P. falciparum (PlasmoDB no. PFL1170w, GenBank accession number EF116593) and human PABP (GenBank accession number CAI 12298). The RNA recognition motifs (RRM) 1–2, 3 and 4 have been highlighted with green, blue and orange color respectively. (b) Three-dimensional models for human PABP and various fragments of PfPABP were created as described in text. The structures have been displayed using the same program as described in legend to . (B) (i)–(iii) Show the modeled structure of human (in yellow) and PfPABP (in green from amino acid 1–180), PfPABP (in blue from amino acid 1–264) and PfPABP (in orange from amino acid 1–531) respectively.
Acknowledgements
The author thanks Moaz Ahmad and Arun Pradhan for help in preparation of figures. The work in R.T. laboratory is supported by Department of Science and Technology grant. Infra-structural support from the Department of Biotechnology, Government of India is gratefully acknowledged.
References
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Ann Rev Biochem 2004; 73:15 - 18
- Preiss T, Hentze MW. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 2003; 25:1201 - 1211
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 2003; 4:223
- Rogers GW, Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A: substrate specificity. J Biol Chem 2001; 276:12598 - 12608
- Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol Cell 2003; 95:157 - 167
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 1999; 68:913 - 963
- Joshi B, Lee K, Maeder DL, Jagus R. Phylogenetic analysis of eIF4E-family members. BMC Evolutionary Biology 2005; 5:48
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol 2000; 20:468 - 477
- Dominguez D, Kislig E, Altmann M, Trachsel H. Structural and functional similarities between the central eukaryotic initiation factor (eIF)4A-binding domain of mammalian eIF4G and the eIF4A-binding domain of yeast eIF4G. Biochem J 2001; 355:223 - 230
- Gregorio ED, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J 1999; 18:4865 - 4874
- De Gaudenzi J, Frasch AC, Clayton C. RNA-binding domain proteins in kinetoplastids: a comparative analysis. Euk Cell 2005; 4:2106 - 2114
- Tuteja R. Malaria-An overview. FEBS J 2007; 274:4670 - 4679
- Pradhan A, Tuteja R. Bipolar, dual Plasmodium falciparum helicase 45 expressed during intraerythrocytic developmental cycle is required for parasite growth. J Mol Biol 2007; 373:268 - 281
- Pradhan A, Hussain ME, Tuteja R. Characterization of replication fork and phosphorylation stimulated Plasmodium falciparum helicase 45. Gene 2008; 420:66 - 75
- Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, et al. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol 1997; 4:717 - 724
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 1997; 89:951 - 961
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, et al. Crystal structures of 7-methylguanosine 5′-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem J 2002; 362:539 - 544
- Tomoo K, Shen X, Okabe K, Nozoe Y, Fukuhara S, Morino S, et al. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J Mol Biol 2003; 328:365 - 383
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 1999; 3:707 - 716
- Bates PA, Sternberg MJE. Model Building by Comparison at CASP3: Using Expert Knowledge and Computer Automation. Proteins: Structure, Function and Genetics Suppl 1999; 3:47 - 54
- Bates PA, Kelley LA, MacCallum RM, Sternberg MJE. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins: Structure, Function and Genetics, Suppl 2001; 5:39 - 46
- Contreras-Moreira B, Bates PA. Domain Fishing: a first step in protein comparative modelling. Bioinformatics 2002; 18:1141 - 1142
- Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell 2001; 7:193 - 203
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet 1995; 11:115 - 116
- Pizzi E, Frontali C. Divergence of noncoding sequences and of insertions encoding non-globular domains at a genomic region well conserved in Plasmodia. J Mol Evol 2000; 50:474 - 480
- Pizzi E, Frontali C. Low-complexity regions in Plasmodium falciparum proteins. Genome Res 2001; 11:218 - 229
- Shaw PJ, Ponmeea N, Karoonuthaisiria N, Kamchonwongpaisana S, Yuthavonga Y. Characterization of human malaria parasite Plasmodium falciparum eIF4E homologue and mRNA 5′ cap status. Mol Biochem Parasitol 2007; 155:146 - 155
- Dhalia R, Reis CRS, Freireb ER, Rochab PO, Katzb R, Munizc JRC, et al. Translation initiation in Leishmania major: characterization of multiple eIF4F subunit homologues. Mol Biochem Parasitol 2005; 140:23 - 41
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 1997; 17:6940 - 6947
- Neff CL, Sachs AB. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol Cell Biol 1999; 19:5557 - 5564
- Tarun SZ Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA 1997; 94:9046 - 9051
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 1998; 17:7480 - 7489
- Gorgona B, Gray NK. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: A developmental perspective. Briefings Functional genomics and proteomic 2004; 3:125 - 141
- Kessler SH, Sachs AB. RNA recognition motif 2 of yeast Pab1p required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol 1998; 18:51 - 57
- Cheng S, Gallie DR. eIF4G, eIFiso4G and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains. J Biol Chem 2007; 282:25247 - 25258