Abstract
The Elmo protein family members are important mediators of small G protein activity, regulating actin-mediated processes such as chemotaxis and engulfment. Until recently,1 Elmo function has not been explored in professional phagocytes such as Dictyostelium discoideum. We discuss the significance of this family with respect to pathways that regulate Rac signaling, we present a comparison of Elmo proteins between representative taxa, and discuss our findings on ElmoA, one of six Elmo proteins found in D. discoideum.
In mammals, leukocytes are the wandering sentinels of the innate immune system that provide the opening gambit against invading parasites. They sense, hunt and kill interlopers of malevolent intent—be it a virus, bacterium or cancer cell—to protect the complex biological system they serve. These fundamental abilities are derived from ancient lineages and are conserved in the social amoeba Dictyostelium discoideum. Over the last several decades, researchers have capitalized on the tractability of D. discoideum as a model system to define the molecular mechanisms that regulate chemotaxis and phagocytosis.Citation2 Generalized, chemotaxis is regulated through pathways that begin with the perception of extracellular signals from target cells or organisms via transmembrane receptors; the signals are then transduced through intracellular networks that function to manipulate changes in the actin-myosin cytoskeleton and induce F-actin formation to direct anterior pseudopod extension and posterior uropod retraction, thus enabling the cell to pursue its target. Dependent on cell-type, the destruction of the target is ultimately achieved through the actin-dependent processes of phagocytosis or granulocyte release.
The small GTPase Rac plays a central role in regulating the actin cytoskeleton to promote cell shape change and movement in many cell types.Citation3,Citation4 Amongst other pathways, extracellular chemoattractant activation of Rac occurs through seven-transmembrane receptors (7-TMRs), which are typically coupled to heterotrimeric G proteins, or through adhesion molecules such as integrins. Rac has been implicated to activate the SCAR (suppressor of cAMP receptor)/WAVE [WASP (Wiskott-Aldrich syndrome protein)family verprolin homology protein] family proteins, which function to activate actin nucleation through the Arp2/3 (actin related protein 2/3) complex.Citation5 SCAR/WAVE is itself found in a regulatory complex that includes Nap1 (Nck-associated protein)/Hem-1 (hematopoietic protein 1), PIR121 (p53-inducible mRNA 121), Abi (Abl-interactor) and HSPC300 (hematopoietic stem progenitor cell 300).Citation6 The exact contribution of the molecules in the complex to SCAR/WAVE activity, whether it is positive, negative or localization- dependent, remains somewhat controversial.Citation6 Early evidence obtained from D. discoideum lacking PIR121 suggested a simple model where the complex functions to negatively regulate SCAR/WAVE activity by sequestrationCitation7,Citation8 until released/activated via Rac-GTP. However, D. discoideum Nap1/Hem-1, among playing a role in other SCAR-independent pathways, was shown to be required for SCAR/WAVE activity.Citation9 Moreover, work in mammalian cells and C. elegans supports a positive role of the complex in the assembly with and activation of Arp2/3.Citation10–Citation12 In addition, elegant work from Weiner and coworkers (2007) using total internal reflection microscopy (TIR-FM) in a live human neutrophil cell line showed that Hem-1/Nap1 localizes to the cell cortex and moves in spiral waves toward the periphery. They observed an inverse relationship between the accumulation of fluorescently-tagged Hem-1/Nap1, and F-actin and with the aid of mathematical modeling, proposed that Hem-1/Nap1 acts as a moving front for actin nucleation via activation the Arp2/3 complex.Citation13
Mounting evidence lead to the broad concept that migrating cells possess a “chemical compass”Citation14 that permits them to “steer” up a shallow extracellular chemoattractant gradient by directing actin polymerization to new pseudopods to better orient the cell. However, Andrew and Insall (2007) have recently challenged this concept with the proposal that directional sensing is fundamentally accomplished by maintaining the most accurate pseudopod. Their evidence from experiments with D. discoideum indicates that new pseudopod generation is stochastic within a specific range of the anterior of the cell, where new pseudopods split from existing ones and are not necessarily generated from the lateral sides of the cell.Citation15,Citation16
Guanine nucleotide exchange factors (GEFs) are the upstream activators of small GTPases like Rac. GEFs function to catalyze the switch of bound GDP (off state) on small G proteins with GTP (on state). GEFs are categorized into two major classes—those that contain the classical Dbl homology-pleckstrin homology domain and those that contain the so-called Docker-ZH2/DHR-2 domain as found in the Dock180 family of proteins. Members of the Dock180 family have been shown to function in conjunction with the Elmo (Engulfment and cell motility) protein family to promote Rac-dependent actin reorganization in both chemotaxis and phagocytosis.Citation17–Citation19 A variety of receptors in organisms ranging from worms to humans can activate the Elmo/Dock180 complex, which include integrins, the PtSer cell death receptor, and 7-TMRs.Citation17 Exactly how the Elmo/Dock180 complex functions in cells has yet to be determined. Some data implicate Elmo to work as a scaffold, with additional roles in localizing the GEF activity performed by Dock180, while other work suggests Elmo is required for Dock180-mediated nucleotide exchange on Rac, where Elmo, nucleotide-free Rac and Dock180 form a ternary structure to execute the addition of GTP.Citation20–Citation22
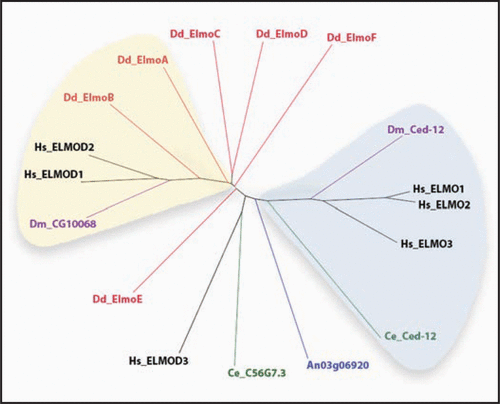
The Elmo family of proteins is conserved throughout evolution. We identified six new members, termed ElmoA–F, encoded by the professional phagocyte D. discoideum. All six proteins contain a centrally located Elmo-domain. Of note are a ∼70 aa C-terminal domain present in ElmoA (see below for significance) that is conserved in three human Elmo proteins (Elmo1–3), an ankyrin repeat region located toward the C-terminus of ElmoD, and the fact that the Elmo domains of ElmoE and F are interrupted with repetitive sequence, a feature typically found in many D. discoideum proteins. Shown in is a relatedness tree of Elmo proteins from several representative species whose genome sequence has been determined. A comparison of the Elmo domains is shown. Excepting the fungus representative Aspergillus niger, all species encode more than one Elmo protein with H. sapiens and D. discoideum each encoding six. No Elmo proteins were observed in yeast. Highlighted in light blue in are human ELMO1–3, and D. melanogaster and C. elegans Ced-12, which have been characterized to form a complex with Dock180 and promote Rac guanine nucleotide exchange to activate actin polymerization. D. discoideum ElmoE and F are most closely related to this grouping, and consistent with its proximity, preliminary characterization of ElmoE function suggests a positive role in actin polymerization (Yan and Jin, unpublished).
Human ElmoD1 and D2 have been recently described as GTPase-activating proteins (GAP) for small G protein ARF family, indicating that Elmo proteins do not follow a simple functional paradigm, having both GEF and GAP promoting activity. Human ElmoD1 and D2 form a strong clade with two uncharacterized Elmo proteins from D. discoideum (ElmoB) and D. melanogaster (CG10068; , yellow highlight). Also proximal to this clade is D. discoideum ElmoA (, yellow highlight). While GAP activity has not been analyzed for ElmoA, we have demonstrated that it negatively regulates actin polymerization—a downstream target of Rac (see details below). Future work on the members of this putative functional clade may reveal an evolutionarily conserved yin and yang regulation of small G proteins by the Elmo family.
Until now, Elmo protein function had not been analyzed in professional phagocytes such as D. discoideum.Citation1 We showed in our first study that ElmoA acts to negatively regulate actin polymerization during phagocytosis and chemotaxis.Citation1 We found that cells lacking ElmoA displayed an increase in phagocytosis, the converse to what was observed for ced12/elmo C. elegans mutants.Citation19 By using biochemical and imaging methodologies, we demonstrated that the overall F-actin level and the secondary F-actin response to chemoattractant stimulation was significantly elevated in elmoA− cells. When we analyzed the relationship between ElmoA and F-actin localization dynamics by TIR-FM in live cells, we measured an inverse correlation between these molecules proximal to the cell cortex. Specifically, we observed that ElmoA would maximally disperse from the cell cortex with a ∼2 sec delay from the peak of an F-actin event; these events were seen either passively or could be induced by the application of a saturating dose of chemoattractant. In addition, the converse was true if we disrupted F-actin in cells with a depolymerizing drug—treatment with Latrunculin B caused ElmoA to accumulate at the cortex as F-actin dissipated. In pull down assays, we found that ElmoA associated with myosin II and actin, and TIR-FM analysis indicated ElmoA dynamics was dependent upon myosin II. Deletion of ∼70 amino acids from the C-terminus revealed a region that was necessary for wild-type function and cortical dispersion upon actin polymerization. While we found that ElmoA did not influence gradient sensing in cells, it did impact the ability of cells to move up a gradient. Loss of elmoA caused excessive pseudopod splitting in chemotaxis assays using broad shallow gradients. ElmoA's apparent role in inhibiting spurious pseudopods fits with the stochastic model of anterior pseudopod splitting proposed by Andrew and Insall (2007).Citation16 In the model, pseudopods are randomly generated and split from a region of the cell containing an existing pseudopod, i.e., there is no propensity for the new pseudopod to form in the direction of the gradient. However, the pseudopod most appropriately aligned toward the gradient has the better chance of being maintained to productively move the cell. The SCAR/WAVE complex is likely involved in regulating these processes as loss of PIR121 affects pseudopod maintenance,Citation7,Citation9,Citation16 and the remarkably similar chemotactic behavior of pirA− and elmoA− null cells suggest that ElmoA may be an upstream negative regulator of SCAR/WAVE activity.
Our data suggests that ElmoA is localized to the actin/myosin II cortex via association with myosin II and functions to inhibit actin polymerization, consistent with a model where the cortex serves to prevent spurious pseudopod formation and to provide mechanical rigidity.Citation7,Citation23 Based on Elmo association with Dock proteins in other systems, we speculate that ElmoA may function to inhibit localized Dock-mediated GEF activity and/or may act as a GAP similar to human ElmoD1 and D2, consistent with its evolutionary proximity to these human proteins (see ). The delayed dispersion kinetics of ElmoA from the cortex upon the build-up of F-actin and the in vivo behavior of the C-terminal ElmoA deletion mutant suggests that F-actin formation, once initiated, can feedback upon ElmoA to promote its dispersion. Dispersion of ElmoA from the cortex, in turn, may free Dock from inhibition or remove localized GAP activity, thus, promoting actin polymerization. Based on the presence of multiple Elmo proteins in D. discoideum, we speculate that chemoattractant stimulation triggers Dock/Elmo-associated GEF activity to promote actin polymerization through the SCAR/WAVE complex. Again, consistent with this idea is our observation that ElmoE co-localizes with F-actin in cells and is most closely related to the Rac-activating Elmo proteins (Yan and Jin T, unpublished; see ).
On a final technical note, our observations of the inverse dynamics of F-actin accumulation and ElmoA dispersion proximal to the plasma membrane would have been missed if we had not visualized fluorescently-tagged proteins in live cells by TIR-FM. TIR-FM is a high-contrast, spatially-limited technique that does not excite fluorescent molecules beyond ∼100 nm from the cover slip. Because interference from potential cytoplasmic fluorescent signal is absent, the technique becomes ideal for the observation of proteins that are imbedded within or reside close to the plasma membrane. In our experiments, the vast proportion of ElmoA-GFP signal was localized to the cytoplasm, making dispersion events unnoticeable in cells co-expressing an RFP-tagged F-actin marker by confocal microscopy. With TIR-FM, we were able to simultaneously visualize and quantify the rise in RFP intensity, signifying the formation of F-actin, and the concomitant, although phase-shifted in time, decrease in ElmoA-GFP intensity in live cells. Interestingly, the advantage of TIR-FM was also capitalized by Weiner and coworkers (2007),Citation13 enabling them to visualize the dynamic spiral wave patterns of the SCAR/WAVE complex protein Hem-1/Nap1 as well as Rac activation in live neutrophil-like cells, events that also could not be observed with standard imaging techniques.
Figures and Tables
Figure 1 A relatedness tree of Elmo domains from proteins of five species is shown. The domain borders (indicated after each protein name below) were identified by a protein blast query of the full-length sequence. Species represented are Dictyostelium discoideum (Dd; red; elmoA: 364–551; elmoB: 106–266; elmoC: 363–547; elmoD: 288–456; elmoE (split domain): 477–566, 628–700; and elmoF (split domain): 259–350, 410–478), Homo sapiens (Hs; black; ELMO1: 300–482; ELMO2: 292–475; ELMO3: 175–356; ELMOD1: 115–304; ELMOD2: 112–272; ELMOD3: 151–314), Caenorhabditis elegans (Ce; green; C56G7.3: 123–293; Ced-12: 362–475), Drosophila melanogaster (Dm; purple; CG10068-PA: 121–290; Ced-12: 297–480) and Aspergillus niger (An; blue; AnCG5336: 220–410). Note that the Dd Elmo domains of elmoE and elmoF are split by poly-glutamine stretches, common in Dictyostelium proteins, which have been eliminated for the alignment. Highlighted in light blue are Elmo proteins reported to promote GEF activity. Highlighted in yellow are Elmo proteins speculated to promote GAP activity based on their proximity to the functionally characterized human ElmoD1 and D2 proteins and on preliminary results. Sequences were aligned with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The resulting guide tree (dnd) was used to create a circular tree in the Java phylogenetic tree viewer Hypertree (http://kinase.com/tools/HyperTree.html; and ref. Citation24).
Acknowledgements
This commentary is supported by NIAID/NIH intramural funds.
Addendum to:
References
- Isik N, Brzostowski JA, Jin T. An Elmo-like protein associated with myosin II restricts spurious F-actin events to coordinate phagocytosis and chemotaxis. Dev Cell 2008; 15:590 - 602
- Jin T, Xu X, Fang J, Isik N, Yan J, Brzostowski JA, Hereld D. How human leukocytes track down and destroy pathogens: lessons learned from the model organism Dictyostelium discoideum. Immunol Res 2009; 43:118 - 127
- Kolsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci 2008; 121:551 - 559
- Dinauer MC. Regulation of neutrophil function by Rac GTPases. Curr Opin Hematol 2003; 10:8 - 15
- Soderling SH, Scott JD. WAVE signalling: from biochemistry to biology. Biochem Soc Trans 2006; 34:73 - 76
- Ibarra N, Pollitt A, Insall RH. Regulation of actin assembly by SCAR/WAVE proteins. Biochem Soc Trans 2005; 33:1243 - 1246
- Blagg SL, Stewart M, Sambles C, Insall RH. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr Biol 2003; 13:1480 - 1487
- Pollitt AY, Blagg SL, Ibarra N, Insall RH. Cell motility and SCAR localisation in axenically growing Dictyostelium cells. Eur J Cell Biol 2006; 85:1091 - 1098
- Ibarra N, Blagg SL, Vazquez F, Insall RH. Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr Biol 2006; 16:717 - 722
- Patel FB, Bernadskaya YY, Chen E, Jobanputra A, Pooladi Z, Freeman KL, et al. The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Dev Biol 2008; 324:297 - 309
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J 2004; 23:749 - 759
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol 2004; 6:319 - 327
- Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol 2007; 5:221
- Bourne HR, Weiner O. A chemical compass. Nature 2002; 419:21
- Insall R, Andrew N. Chemotaxis in Dictyostelium: how to walk straight using parallel pathways. Curr Opin Microbiol 2007; 10:578 - 581
- Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol 2007; 9:193 - 200
- Cote JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol 2007; 17:383 - 393
- Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol 2007; 7:964 - 974
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol 2004; 20:193 - 221
- Lu M, Kinchen JM, Rossman KL, Grimsley C, Hall M, Sondek J, et al. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr Biol 2005; 15:371 - 377
- Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol 2002; 4:574 - 582
- Grimsley CM, Kinchen JM, Tosello-Trampont AC, Brugnera E, Haney LB, Lu M, et al. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Racdependent cell migration. J Biol Chem 2004; 279:6087 - 6097
- Zhang H, Wessels D, Fey P, Daniels K, Chisholm RL, Soll DR. Phosphorylation of the myosin regulatory light chain plays a role in motility and polarity during Dictyostelium chemotaxis. J Cell Sci 2002; 115:1733 - 1747
- Bingham J, Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics 2000; 16:660 - 661