Abstract
Regulation of alternative splicing is coupled to transcription quality, the polymerase elongation rate being an important factor in modulating splicing choices. In a recently published work, we provide evidence that intragenic histone acetylation patterns can be affected by neural cell excitation in order to regulate alternative splicing of the neural cell adhesion molecule (NCAM) mRNA. This example illustrates how an extracellular stimulus can influence transcription-coupled alternative splicing, strengthening the link between chromatin structure, transcriptional elongation and mRNA processing.
Alternative splicing is known to be the main contributor to the expansion of proteome expression potentials in metazoans.Citation1 The number of protein variants generated by alternative splicing is particularly high in the nervous system, affecting proteins such as ion channels, cell adhesion molecules, components of the cytoskeleton, proteins involved in signaling and trafficking, transcription factors and even splicing regulators.Citation2–Citation4 The alternative variants have usually different functions and regulatory features, which in some cases may even be antagonistic. Furthermore, since the process is tightly regulated, alternative splicing provides a whole new level for fine control of gene expression, in addition to the extensively studied regulation of transcription initiation. The cellular functions where this mechanism plays a role in neurons include nearly all the aspects of both developing and mature cells.
Cellular components of the nervous system require complex functional regulation, with responses varying in different neuronal types, during development and in response to different stimuli. Given the expansion of functions of alternative splicing in neuronal cells, the precise control of the splicing choices is expected to be of crucial importance for proper cellular functionality. Combinatorial control, including additive and cooperative interactions between different regulatory components, is believed to be responsible for such fine tuning of alternative splicing.Citation4,Citation5 It implies that a change in the activity of a single splicing regulator can affect splicing of specific mRNAs to different degrees in the same cell or influence alternative splicing of one mRNA in one cell type and not in others, depending on the action of other regulatory elements.Citation6,Citation7 In extreme cases, malfunction of the complex network of alternative splicing regulation, and the consequent alterations in splicing patterns, is associated with severe neurological disorders such as ataxia, frontotemporal dementia, myotonic dystrophy and spinal muscular atrophy.Citation8
Transcription and mRNA processing are part of a growing list of nuclear and cytoplasmic mRNA-protein biogenesis steps that need to be taken into account in order to describe gene expression in an integrative way.Citation4,Citation9 Most importantly, these steps are usually coupled, influence each other and can be co-regulated in a concerted way to achieve specific cellular responses.Citation10–Citation12 In particular, the notion that eukaryotic mRNAs are first fully transcribed and then processed is now abandoned, since most mRNA processing steps are known to occur co-transcriptionally in vivo.Citation10,Citation13,Citation14
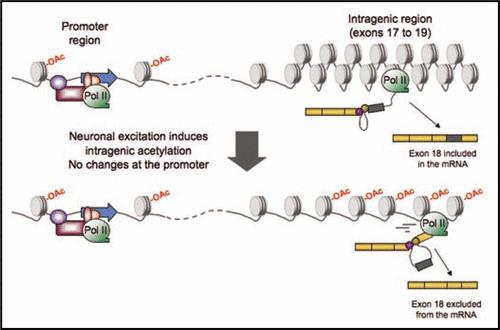
In a recent report,Citation15 we characterized how a dynamical modulation of transcription quality can regulate alternative splicing of an endogenous gene in response to an external cue, such as neural cell excitation. We determined that exon 18 of the gene coding for the neural cell adhesion molecule (NCAM) is preferentially included in the mature mRNA when a slow RNA polymerase II (pol II) mutant is used to drive transcription. This exon is also regulated in response to excitation of neuroblastoma cells and hippocampal neurons: upon cell membrane depolarization a switch is detected from the NCAM 180 isoform mRNA (typical of mature neurons and stable synapses) to the NCAM 140 isoform (typical of developing neurons).Citation15 The dynamic balance between these two isoforms seems to be important for synapses building and remodeling in response to transient activity.Citation16,Citation17 But perhaps the most interesting finding of this work is that the regulatory mechanism involves an excitation-induced modulation of intragenic chromatin, which increases pol II processivity and favors exon 18 skipping.Citation15 Using ChIP and chromatin accessibility assays, we found increased levels of H3K9 acetylation and a concomitant relaxation of the chromatin in the region comprised between exons 17 and the 3′ end of the gene. Surprisingly, none of these changes affect the gene promoter region, suggesting that transcription initiation is not affected. Finally, the effect of depolarization on splicing can be both enhanced and mimicked using the HDAC inhibitor trichostatin A, further implicating histone acetylation in the regulation of this alternative splicing event.Citation15
Our group has been investigating for over a decade the coupling between transcription and alternative splicing regulation, since the discovery that different promoters specify different alternative splicing patterns, and that this phenomenon is related more to transcription quality than quantity.Citation18 Traditionally, alternative splicing events are thought to be subjected to the influence of neighboring elements that regulate splicing choices, typically short cis-acting sequences which can be in the exons or adjacent introns. These sequences serve as docking sites for trans-acting regulatory proteins that help or obstruct splice site recognition and bridging.Citation19 In addition, the pre-mRNA secondary structure might play important roles, hiding or helping to present splice sites and regulatory sequences to the nuclear processing machinery.Citation20 The work of our group and others has resulted in a large body of evidence that supports a very intuitive notion that complements this view: the rate of transcriptional elongation along the usually long metazoan genes contributes to alternative splicing regulation.Citation21,Citation22
Since pre-mRNA is synthesized roughly at the same time as it is being recognized by the processing machinery, the function of all the mentioned regulatory elements can be affected by the “transcriptional speed”. First, recognition of weak splice sites would be favored if the transcription of competing ones is delayed. Also, the timing of transcription of splicing enhancers or silencers could determine their effective influence on splicing regulation. The same would be true for relevant RNA secondary structures. Finally, since many splicing regulatory proteins are thought to be recruited by the pol II machinery itself, the quality of transcription would affect the action of these trans-acting regulators as well.
The NCAM model, shown in , illustrates one of the ways in which transcription quality can be modulated in vivo, namely modifying the chromatin structure of the transcription template. An alternative way to achieve this would be the modification of the pol II machinery itself. In agreement with the first option, it has been reported that in certain conditions chromatin remodeling complexes can create roadblocks to transcription that also modulate alternative splicing in the CD44 gene.Citation23 Alteration of different features of chromatin structure is a perfect way for integrating several pathways in the transcriptional regulation, a matter whose importance is widely established for the nervous system.Citation24,Citation25 In particular, histone acetylation is known to be crucial for many processes that rely on neuronal excitation, learning being a clear example, with a well supported role of histone acetyl-transferases (HATs) like CBP.Citation26,Citation27 The molecular mechanism that mediates the acetylation changes observed in the NCAM regulation remains to be deciphered. Balance between HAT and HDAC activities will affect the acetylation patterns observed, but how they can be recruited differentially to intragenic region is a matter that has not been explored in detail yet.
Evidence indicating that both of these activities can be associated with the RNA pol II can provide a clue.Citation28 Since histone modifications posed by the transcription machinery recruits splicing factors,Citation29 and alternative splicing factors such as the SR protein SC35 can regulate pol II elongation,Citation30 the elucidation of the interplay between chromatin, elongation and mRNA processing seems a complex and exciting task to be undertaken. In this direction, the regulation of intragenic histone acetylation patterns is claiming for attention as an actor in this play.
Figures and Tables
Figure 1 Model of neuronal excitation-induced regulation of alternative splicing for the NCAM exón 18. Intragenic histones located between exons 17 and 19 are basally hipo-acetylated, while chromatin adopts a closed structure (top). Upon depolarization with high extracelular KCl concentration, histone acetyaltion and chromatin relaxation is detected in this región (bottom). Concomitantly, RNA pol II processivity increases, a situation which is associated with alternative exon exclusion in co-transcriptional splicing since less time is available for recognition of the weak exon (exon 18, which is shown in dark gray). The transcriptional changes affect only elongation, since no changes are detected at the promoter región (marked with a blue arrow).
Addendum to:
References
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002; 418:236 - 243
- Fagnani M, Barash Y, Ip JY, Misquitta C, Pan Q, Saltzman AL, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol 2007; 8:108
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci 2007; 8:819 - 831
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol 2006; 16:102 - 110
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 2000; 25:381 - 388
- Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol 2005; 3:158
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 2005; 37:844 - 852
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron 2006; 52:93 - 101
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell 2002; 108:439 - 451
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 2005; 17:251 - 256
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009; 136:688 - 700
- Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol 2005; 12:1037 - 1044
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 2005; 19:65 - 75
- Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 2006; 13:815 - 822
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA 2009; 106:4325 - 4330
- Ronn LC, Berezin V, Bock E. The neural cell adhesion molecule in synaptic plasticity and ageing. Int J Dev Neurosci 2000; 18:193 - 199
- Hoffman KB, Murray BA, Lynch G, Munirathinam S, Bahr BA. Delayed and isoformspecific effect of NMDA exposure on neural cell adhesion molecules in hippocampus. Neurosci Res 2001; 39:167 - 173
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA 1997; 94:11456 - 11460
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 2003; 72:291 - 336
- Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 2004; 24:10505 - 10514
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 2003; 12:525 - 532
- Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol 2006; 13:5 - 7
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 2006; 13:22 - 29
- Colvis CM, Pollock JD, Goodman RH, Impey S, Dunn J, Mandel G, et al. Epigenetic mechanisms and gene networks in the nervous system. J Neurosci 2005; 25:10379 - 10389
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci 2005; 6:108 - 118
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, et al. Chromatin acetylation, memory and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 2004; 42:947 - 959
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 2004; 42:961 - 972
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707 - 719
- Sims RJ 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 2007; 28:665 - 676
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 2008; 15:819 - 826